siRNA that participates in Drosophila dosage compensation is produced by many 1.688X and 359 bp repeats
- PMID: 38718207
- PMCID: PMC11228850
- DOI: 10.1093/genetics/iyae074
siRNA that participates in Drosophila dosage compensation is produced by many 1.688X and 359 bp repeats
Abstract
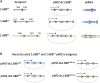
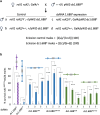
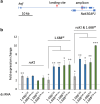
Organisms with differentiated sex chromosomes must accommodate unequal gene dosage in males and females. Male fruit flies increase X-linked gene expression to compensate for hemizygosity of their single X chromosome. Full compensation requires localization of the Male-Specific Lethal (MSL) complex to active genes on the male X, where it modulates chromatin to elevate expression. The mechanisms that identify X chromatin are poorly understood. The euchromatic X is enriched for AT-rich, ∼359 bp satellites termed the 1.688X repeats. Autosomal insertions of 1.688X DNA enable MSL recruitment to nearby genes. Ectopic expression of dsRNA from one of these repeats produces siRNA and partially restores X-localization of MSLs in males with defective X recognition. Surprisingly, expression of double-stranded RNA from three other 1.688X repeats failed to rescue males. We reconstructed dsRNA-expressing transgenes with sequence from two of these repeats and identified phasing of repeat DNA, rather than sequence or orientation, as the factor that determines rescue of males with defective X recognition. Small RNA sequencing revealed that siRNA was produced in flies with a transgene that rescues, but not in those carrying a transgene with the same repeat but different phasing. We demonstrate that pericentromeric X heterochromatin promotes X recognition through a maternal effect, potentially mediated by small RNA from closely related heterochromatic repeats. This suggests that the sources of siRNAs promoting X recognition are highly redundant. We propose that enrichment of satellite repeats on Drosophilid X chromosomes facilitates the rapid evolution of differentiated sex chromosomes by marking the X for compensation.
Keywords: Drosophila melanogaster; siRNA; roX; 1.688X satellite repeats; 359 bp repeats; X recognition; dosage compensation; epigenetics; small RNA.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures


References
-
- Alekseyenko AA, Ellison CE, Gorchakov AA, Zhou Q, Kaiser VB, Toda N, Walton Z, Peng S, Park PJ, Bachtrog D, et al. 2013. Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev. 27(8):853–858. doi:10.1101/gad.215426.113. - DOI - PMC - PubMed
-
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 134(4):599–609. doi:10.1016/j.cell.2008.06.033. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

