Phosphorylation in the Ser/Arg-rich region of the nucleocapsid of SARS-CoV-2 regulates phase separation by inhibiting self-association of a distant helix
- PMID: 38718862
- PMCID: PMC11180338
- DOI: 10.1016/j.jbc.2024.107354
Phosphorylation in the Ser/Arg-rich region of the nucleocapsid of SARS-CoV-2 regulates phase separation by inhibiting self-association of a distant helix
Abstract
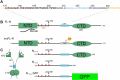
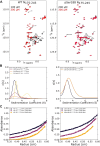
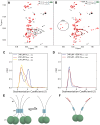
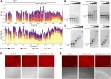
The nucleocapsid protein (N) of SARS-CoV-2 is essential for virus replication, genome packaging, evading host immunity, and virus maturation. N is a multidomain protein composed of an independently folded monomeric N-terminal domain that is the primary site for RNA binding and a dimeric C-terminal domain that is essential for efficient phase separation and condensate formation with RNA. The domains are separated by a disordered Ser/Arg-rich region preceding a self-associating Leu-rich helix. Phosphorylation in the Ser/Arg region in infected cells decreases the viscosity of N:RNA condensates promoting viral replication and host immune evasion. The molecular level effect of phosphorylation, however, is missing from our current understanding. Using NMR spectroscopy and analytical ultracentrifugation, we show that phosphorylation destabilizes the self-associating Leu-rich helix 30 amino-acids distant from the phosphorylation site. NMR and gel shift assays demonstrate that RNA binding by the linker is dampened by phosphorylation, whereas RNA binding to the full-length protein is not significantly affected presumably due to retained strong interactions with the primary RNA-binding domain. Introducing a switchable self-associating domain to replace the Leu-rich helix confirms the importance of linker self-association to droplet formation and suggests that phosphorylation not only increases solubility of the positively charged elongated Ser/Arg region as observed in other RNA-binding proteins but can also inhibit self-association of the Leu-rich helix. These data highlight the effect of phosphorylation both at local sites and at a distant self-associating hydrophobic helix in regulating liquid-liquid phase separation of the entire protein.
Keywords: AUC; LLPS; NMR; SARS-CoV-2; phosphorylation; protein RNA interactions.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interests The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein.Nat Commun. 2021 Jan 21;12(1):502. doi: 10.1038/s41467-020-20768-y. Nat Commun. 2021. PMID: 33479198 Free PMC article.
-
ATP biphasically modulates LLPS of SARS-CoV-2 nucleocapsid protein and specifically binds its RNA-binding domain.Biochem Biophys Res Commun. 2021 Feb 19;541:50-55. doi: 10.1016/j.bbrc.2021.01.018. Epub 2021 Jan 14. Biochem Biophys Res Commun. 2021. PMID: 33477032 Free PMC article.
-
Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions.Mol Cell. 2020 Dec 17;80(6):1092-1103.e4. doi: 10.1016/j.molcel.2020.11.025. Epub 2020 Nov 20. Mol Cell. 2020. PMID: 33248025 Free PMC article.
-
Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization.J Biol Chem. 2024 Nov;300(11):107834. doi: 10.1016/j.jbc.2024.107834. Epub 2024 Sep 27. J Biol Chem. 2024. PMID: 39343000 Free PMC article. Review.
-
Phase separation by the SARS-CoV-2 nucleocapsid protein: Consensus and open questions.J Biol Chem. 2022 Mar;298(3):101677. doi: 10.1016/j.jbc.2022.101677. Epub 2022 Feb 4. J Biol Chem. 2022. PMID: 35131265 Free PMC article. Review.
Cited by
-
Intrinsic factors behind long COVID: exploring the role of nucleocapsid protein in thrombosis.PeerJ. 2025 May 20;13:e19429. doi: 10.7717/peerj.19429. eCollection 2025. PeerJ. 2025. PMID: 40416618 Free PMC article. Review.
-
The Nucleocapsid (N) Proteins of Different Human Coronaviruses Demonstrate a Variable Capacity to Induce the Formation of Cytoplasmic Condensates.Int J Mol Sci. 2024 Dec 7;25(23):13162. doi: 10.3390/ijms252313162. Int J Mol Sci. 2024. PMID: 39684875 Free PMC article.
-
Afobazole: a potential drug candidate which can inhibit SARS CoV-2 and mimicry of the human respiratory pacemaker protein.In Silico Pharmacol. 2025 Feb 17;13(1):30. doi: 10.1007/s40203-025-00316-6. eCollection 2025. In Silico Pharmacol. 2025. PMID: 39974371
-
Phosphorylation Toggles the SARS-CoV-2 Nucleocapsid Protein Between Two Membrane-Associated Condensate States.bioRxiv [Preprint]. 2025 Jun 10:2024.10.17.618867. doi: 10.1101/2024.10.17.618867. bioRxiv. 2025. PMID: 39464032 Free PMC article. Preprint.
-
Dynamic interactions of dimeric hub proteins underlie their diverse functions and structures: A comparative analysis of 14-3-3 and LC8.J Biol Chem. 2025 Apr;301(4):108416. doi: 10.1016/j.jbc.2025.108416. Epub 2025 Mar 17. J Biol Chem. 2025. PMID: 40107617 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

