STAT3 activation of SCAP-SREBP-1 signaling upregulates fatty acid synthesis to promote tumor growth
- PMID: 38718868
- PMCID: PMC11176798
- DOI: 10.1016/j.jbc.2024.107351
STAT3 activation of SCAP-SREBP-1 signaling upregulates fatty acid synthesis to promote tumor growth
Abstract
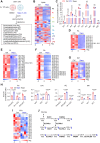
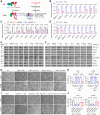
SCAP plays a central role in controlling lipid homeostasis by activating SREBP-1, a master transcription factor in controlling fatty acid (FA) synthesis. However, how SCAP expression is regulated in human cancer cells remains unknown. Here, we revealed that STAT3 binds to the promoter of SCAP to activate its expression across multiple cancer cell types. Moreover, we identified that STAT3 also concurrently interacts with the promoter of SREBF1 gene (encoding SREBP-1), amplifying its expression. This dual action by STAT3 collaboratively heightens FA synthesis. Pharmacological inhibition of STAT3 significantly reduces the levels of unsaturated FAs and phospholipids bearing unsaturated FA chains by reducing the SCAP-SREBP-1 signaling axis and its downstream effector SCD1. Examination of clinical samples from patients with glioblastoma, the most lethal brain tumor, demonstrates a substantial co-expression of STAT3, SCAP, SREBP-1, and SCD1. These findings unveil STAT3 directly regulates the expression of SCAP and SREBP-1 to promote FA synthesis, ultimately fueling tumor progression.
Keywords: FASN; SCAP; SCD1; SREBP-1; STAT3; fatty acid; glioblastoma; lipogenesis; phospholipid.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

