Comprehensive single-cell analysis deciphered microenvironmental dynamics and immune regulator olfactomedin 4 in pathogenesis of gallbladder cancer
- PMID: 38719336
- PMCID: PMC11347255
- DOI: 10.1136/gutjnl-2023-331773
Comprehensive single-cell analysis deciphered microenvironmental dynamics and immune regulator olfactomedin 4 in pathogenesis of gallbladder cancer
Abstract
Objective: Elucidating complex ecosystems and molecular features of gallbladder cancer (GBC) and benign gallbladder diseases is pivotal to proactive cancer prevention and optimal therapeutic intervention.
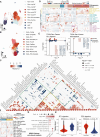
Design: We performed single-cell transcriptome analysis on 230 737 cells from 15 GBCs, 4 cholecystitis samples, 3 gallbladder polyps, 5 gallbladder adenomas and 16 adjacent normal tissues. Findings were validated through large-scale histological assays, digital spatial profiler multiplexed immunofluorescence (GeoMx), etc. Further molecular mechanism was demonstrated with in vitro and in vivo studies.
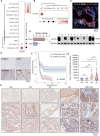
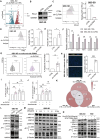
Results: The cell atlas unveiled an altered immune landscape across different pathological states of gallbladder diseases. GBC featured a more suppressive immune microenvironment with distinct T-cell proliferation patterns and macrophage attributions in different GBC subtypes. Notably, mutual exclusivity between stromal and immune cells was identified and remarkable stromal ecosystem (SC) heterogeneity during GBC progression was unveiled. Specifically, SC1 demonstrated active interaction between Fibro-iCAF and Endo-Tip cells, correlating with poor prognosis. Moreover, epithelium genetic variations within adenocarcinoma (AC) indicated an evolutionary similarity between adenoma and AC. Importantly, our study identified elevated olfactomedin 4 (OLFM4) in epithelial cells as a central player in GBC progression. OLFM4 was related to T-cell malfunction and tumour-associated macrophage infiltration, leading to a worse prognosis in GBC. Further investigations revealed that OLFM4 upregulated programmed death-ligand 1 (PD-L1) expression through the MAPK-AP1 axis, facilitating tumour cell immune evasion.
Conclusion: These findings offer a valuable resource for understanding the pathogenesis of gallbladder diseases and indicate OLFM4 as a potential biomarker and therapeutic target for GBC.
Keywords: adenoma; gallbladder cancer; inflammation; polyp.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
-
- Javle MM, Hainsworth JD, Swanton C, et al. . Pertuzumab + Trastuzumab for Her2-positive metastatic biliary cancer: preliminary data from Mypathway. JCO 2017;35:402. 10.1200/JCO.2017.35.4_suppl.402 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
