Combinatorial microRNA activity is essential for the transition of pluripotent cells from proliferation into dormancy
- PMID: 38719471
- PMCID: PMC11146600
- DOI: 10.1101/gr.278662.123
Combinatorial microRNA activity is essential for the transition of pluripotent cells from proliferation into dormancy
Abstract
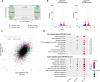
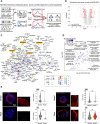
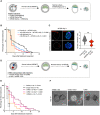
Dormancy is a key feature of stem cell function in adult tissues as well as in embryonic cells in the context of diapause. The establishment of dormancy is an active process that involves extensive transcriptional, epigenetic, and metabolic rewiring. How these processes are coordinated to successfully transition cells to the resting dormant state remains unclear. Here we show that microRNA activity, which is otherwise dispensable for preimplantation development, is essential for the adaptation of early mouse embryos to the dormant state of diapause. In particular, the pluripotent epiblast depends on miRNA activity, the absence of which results in the loss of pluripotent cells. Through the integration of high-sensitivity small RNA expression profiling of individual embryos and protein expression of miRNA targets with public data of protein-protein interactions, we constructed the miRNA-mediated regulatory network of mouse early embryos specific to diapause. We find that individual miRNAs contribute to the combinatorial regulation by the network, and the perturbation of the network compromises embryo survival in diapause. We further identified the nutrient-sensitive transcription factor TFE3 as an upstream regulator of diapause-specific miRNAs, linking cytoplasmic MTOR activity to nuclear miRNA biogenesis. Our results place miRNAs as a critical regulatory layer for the molecular rewiring of early embryos to establish dormancy.
© 2024 Iyer et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
