MeCP2 binds to methylated DNA independently of phase separation and heterochromatin organisation
- PMID: 38719804
- PMCID: PMC11079052
- DOI: 10.1038/s41467-024-47395-1
MeCP2 binds to methylated DNA independently of phase separation and heterochromatin organisation
Abstract
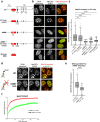
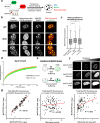
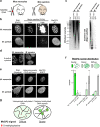
Correlative evidence has suggested that the methyl-CpG-binding protein MeCP2 contributes to the formation of heterochromatin condensates via liquid-liquid phase separation. This interpretation has been reinforced by the observation that heterochromatin, DNA methylation and MeCP2 co-localise within prominent foci in mouse cells. The findings presented here revise this view. MeCP2 localisation is independent of heterochromatin as MeCP2 foci persist even when heterochromatin organisation is disrupted. Additionally, MeCP2 foci fail to show hallmarks of phase separation in live cells. Importantly, we find that mouse cellular models are highly atypical as MeCP2 distribution is diffuse in most mammalian species, including humans. Notably, MeCP2 foci are absent in Mus spretus which is a mouse subspecies lacking methylated satellite DNA repeats. We conclude that MeCP2 has no intrinsic tendency to form condensates and its localisation is independent of heterochromatin. Instead, the distribution of MeCP2 in the nucleus is primarily determined by global DNA methylation patterns.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

