Development of a new affinity maturation protocol for the construction of an internalizing anti-nucleolin antibody library
- PMID: 38719911
- PMCID: PMC11079059
- DOI: 10.1038/s41598-024-61230-z
Development of a new affinity maturation protocol for the construction of an internalizing anti-nucleolin antibody library
Abstract
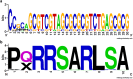
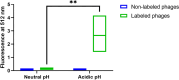
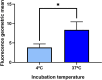
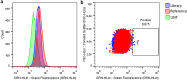
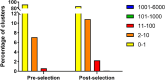
Over the last decades, monoclonal antibodies have substantially improved the treatment of several conditions. The continuous search for novel therapeutic targets and improvements in antibody's structure, demands for a constant optimization of their development. In this regard, modulation of an antibody's affinity to its target has been largely explored and culminated in the discovery and optimization of a variety of molecules. It involves the creation of antibody libraries and selection against the target of interest. In this work, we aimed at developing a novel protocol to be used for the affinity maturation of an antibody previously developed by our group. An antibody library was constructed using an in vivo random mutagenesis approach that, to our knowledge, has not been used before for antibody development. Then, a cell-based phage display selection protocol was designed to allow the fast and simple screening of antibody clones capable of being internalized by target cells. Next generation sequencing coupled with computer analysis provided an extensive characterization of the created library and post-selection pool, that can be used as a guide for future antibody development. With a single selection step, an enrichment in the mutated antibody library, given by a decrease in almost 50% in sequence diversity, was achieved, and structural information useful in the study of the antibody-target interaction in the future was obtained.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Rapid Affinity Maturation of Novel Anti-PD-L1 Antibodies by a Fast Drop of the Antigen Concentration and FACS Selection of Yeast Libraries.Biomed Res Int. 2019 Dec 28;2019:6051870. doi: 10.1155/2019/6051870. eCollection 2019. Biomed Res Int. 2019. PMID: 31976323 Free PMC article.
-
Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation.Int J Mol Sci. 2019 Apr 15;20(8):1861. doi: 10.3390/ijms20081861. Int J Mol Sci. 2019. PMID: 30991723 Free PMC article. Review.
-
A novel lentiviral scFv display library for rapid optimization and selection of high affinity antibodies.Biochem Biophys Res Commun. 2018 Apr 30;499(1):71-77. doi: 10.1016/j.bbrc.2018.03.131. Epub 2018 Mar 20. Biochem Biophys Res Commun. 2018. PMID: 29559238
-
Design and construction of a naïve mouse antibody phage display library.J Immunol Methods. 2010 Feb 28;353(1-2):31-43. doi: 10.1016/j.jim.2010.01.003. Epub 2010 Jan 18. J Immunol Methods. 2010. PMID: 20093119
-
A review of in vitro stochastic and non-stochastic affinity maturation strategies for phage display derived monoclonal antibodies.Int J Biol Macromol. 2024 Oct;277(Pt 2):134217. doi: 10.1016/j.ijbiomac.2024.134217. Epub 2024 Jul 26. Int J Biol Macromol. 2024. PMID: 39069045 Review.
Cited by
-
Development of FAP-targeted theranostics discovered by next-generation sequencing-augmented mining of a novel immunized VNAR library.bioRxiv [Preprint]. 2025 Jan 13:2025.01.13.632555. doi: 10.1101/2025.01.13.632555. bioRxiv. 2025. PMID: 39868181 Free PMC article. Preprint.
-
Monoclonal antibodies: From magic bullet to precision weapon.Mol Biomed. 2024 Oct 11;5(1):47. doi: 10.1186/s43556-024-00210-1. Mol Biomed. 2024. PMID: 39390211 Free PMC article. Review.
-
The characterization of variable new antigen receptors targeting FAP isolated from a novel immunized library.Commun Biol. 2025 Aug 13;8(1):1210. doi: 10.1038/s42003-025-08610-x. Commun Biol. 2025. PMID: 40804296 Free PMC article.
References
-
- Roskos L, Klakamp S, Liang M, Arends R, Green L. Antibody affinity. In: Dübel S, Reichert JM, editors. Handbook of therapeutic antibodies. Wiley; 2014. pp. 115–139.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

