Recycled melanoma-secreted melanosomes regulate tumor-associated macrophage diversification
- PMID: 38719996
- PMCID: PMC11377571
- DOI: 10.1038/s44318-024-00103-7
Recycled melanoma-secreted melanosomes regulate tumor-associated macrophage diversification
Abstract
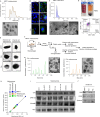
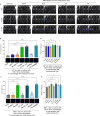
Extracellular vesicles (EVs) are important mediators of communication between cells. Here, we reveal a new mode of intercellular communication by melanosomes, large EVs secreted by melanocytes for melanin transport. Unlike small EVs, which are disintegrated within the receiver cell, melanosomes stay intact within them, gain a unique protein signature, and can then be further transferred to another cell as "second-hand" EVs. We show that melanoma-secreted melanosomes passaged through epidermal keratinocytes or dermal fibroblasts can be further engulfed by resident macrophages. This process leads to macrophage polarization into pro-tumor or pro-immune cell infiltration phenotypes. Melanosomes that are transferred through fibroblasts can carry AKT1, which induces VEGF secretion from macrophages in an mTOR-dependent manner, promoting angiogenesis and metastasis in vivo. In melanoma patients, macrophages that are co-localized with AKT1 are correlated with disease aggressiveness, and immunotherapy non-responders are enriched in macrophages containing melanosome markers. Our findings suggest that interactions mediated by second-hand extracellular vesicles contribute to the formation of the metastatic niche, and that blocking the melanosome cues of macrophage diversification could be helpful in halting melanoma progression.
Keywords: Angiogenesis; Cell-to-Cell-Transfer; Heterogeneity; Melanosomes; Tumor Associated Macrophages.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Ando H, Yoshimoto S, Yoshida M, Shimoda N, Tadokoro R, Kohda H, Ishikawa M, Nishikata T, Katayama B, Ozawa T et al (2020) Dermal fibroblasts internalize phosphatidylserine-exposed secretory melanosome clusters and apoptotic melanocytes. Int J Mol Sci 21:5789 10.3390/ijms21165789 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

