Refined analytical pipeline for the pharmacodynamic assessment of T-cell responses to vaccine antigens
- PMID: 38720900
- PMCID: PMC11076743
- DOI: 10.3389/fimmu.2024.1404121
Refined analytical pipeline for the pharmacodynamic assessment of T-cell responses to vaccine antigens
Abstract
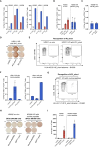
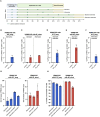
Pharmacodynamic assessment of T-cell-based cancer immunotherapies often focus on detecting rare circulating T-cell populations. The therapy-induced immune cells in blood-derived clinical samples are often present in very low frequencies and with the currently available T-cell analytical assays, amplification of the cells of interest prior to analysis is often required. Current approaches aiming to enrich antigen-specific T cells from human Peripheral Blood Mononuclear Cells (PBMCs) depend on in vitro culturing in presence of their cognate peptides and cytokines. In the present work, we improved a standard, publicly available protocol for T-cell immune analyses based on the in vitro expansion of T cells. We used PBMCs from healthy subjects and well-described viral antigens as a model system for optimizing the experimental procedures and conditions. Using the standard protocol, we first demonstrated significant enrichment of antigen-specific T cells, even when their starting frequency ex vivo was low. Importantly, this amplification occurred with high specificity, with no or neglectable enrichment of irrelevant T-cell clones being observed in the cultures. Testing of modified culturing timelines suggested that the protocol can be adjusted accordingly to allow for greater cell yield with strong preservation of the functionality of antigen-specific T cells. Overall, our work has led to the refinement of a standard protocol for in vitro stimulation of antigen-specific T cells and highlighted its reliability and reproducibility. We envision that the optimized protocol could be applied for longitudinal monitoring of rare blood-circulating T cells in scenarios with limited sample material.
Keywords: T-cell expansion; T-cell response; antigen specificity; cell culture; immune monitoring; in vitro stimulation; pipeline; vaccine antigen.
Copyright © 2024 Pavlidis, Viborg, Lausen, Rønø and Kleine-Kohlbrecher.
Conflict of interest statement
Authors MP, NV, ML, BR and DK-K were employed by the company Evaxion Biotech.
Figures

Similar articles
-
Interferon-gamma production in response to in vitro stimulation with collagen type II in rheumatoid arthritis is associated with HLA-DRB1(*)0401 and HLA-DQ8.Arthritis Res. 2000;2(1):75-84. doi: 10.1186/ar71. Arthritis Res. 2000. PMID: 11219392 Free PMC article.
-
gp100(209-2M) peptide immunization of human lymphocyte antigen-A2+ stage I-III melanoma patients induces significant increase in antigen-specific effector and long-term memory CD8+ T cells.Clin Cancer Res. 2004 Jan 15;10(2):668-80. doi: 10.1158/1078-0432.ccr-0095-03. Clin Cancer Res. 2004. PMID: 14760090
-
High-density preculture of PBMCs restores defective sensitivity of circulating CD8 T cells to virus- and tumor-derived antigens.Blood. 2015 Jul 9;126(2):185-94. doi: 10.1182/blood-2015-01-622704. Epub 2015 May 29. Blood. 2015. PMID: 26024876
-
Melanoma vaccines: the paradox of T cell activation without clinical response.Cancer Chemother Pharmacol. 2000;46 Suppl:S62-6. doi: 10.1007/pl00014052. Cancer Chemother Pharmacol. 2000. PMID: 10950150 Review.
-
T-cell-directed cancer vaccines: the melanoma model.Expert Opin Biol Ther. 2001 Mar;1(2):277-90. doi: 10.1517/14712598.1.2.277. Expert Opin Biol Ther. 2001. PMID: 11727535 Review.
References
-
- Bjoern J, Iversen TZ, Nitschke NJ, Andersen MH, Svane IM. Safety, immune and clinical responses in metastatic melanoma patients vaccinated with a long peptide derived from indoleamine 2,3-dioxygenase in combination with ipilimumab. Cytotherapy. (2016) 18:1043–55. doi: 10.1016/j.jcyt.2016.05.010 - DOI - PubMed
-
- Moderna and Merck Announce mRNA-4157/V940, an Investigational Personalized mRNA Cancer Vaccine, in Combination with KEYTRUDA(R) (pembrolizumab), Met Primary Efficacy Endpoint in Phase 2b KEYNOTE-942 Trial. Available online at: https://investors.modernatx.com/news/news-details/2022/Moderna-and-Merck....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

