Catalytic mechanism and kinetics of malate dehydrogenase
- PMID: 38721782
- PMCID: PMC11461317
- DOI: 10.1042/EBC20230086
Catalytic mechanism and kinetics of malate dehydrogenase
Abstract
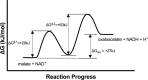
Malate dehydrogenase (MDH) is a ubiquitous and central enzyme in cellular metabolism, found in all kingdoms of life, where it plays vital roles in the cytoplasm and various organelles. It catalyzes the reversible NAD+-dependent reduction of L-malate to oxaloacetate. This review describes the reaction mechanism for MDH and the effects of mutations in and around the active site on catalytic activity and substrate specificity, with a particular focus on the loop that encloses the active site after the substrates have bound. While MDH exhibits selectivity for its preferred substrates, mutations can alter the specificity of MDH for each cosubstrate. The kinetic characteristics and similarities of a variety of MDH isozymes are summarized, and they illustrate that the KM values are consistent with the relative concentrations of the substrates in cells. As a result of its existence in different cellular environments, MDH properties vary, making it an attractive model enzyme for studying enzyme activity and structure under different conditions.
Keywords: enzyme kinetics; enzyme specificity; malate dehydrogenase; reaction mechanism.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

