RSG1 is required for cilia-dependent neural tube closure
- PMID: 38721990
- PMCID: PMC11141724
- DOI: 10.1002/dvg.23602
RSG1 is required for cilia-dependent neural tube closure
Abstract
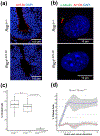
Cilia play a key role in the regulation of signaling pathways required for embryonic development, including the proper formation of the neural tube, the precursor to the brain and spinal cord. Forward genetic screens were used to generate mouse lines that display neural tube defects (NTD) and secondary phenotypes useful in interrogating function. We describe here the L3P mutant line that displays phenotypes of disrupted Sonic hedgehog signaling and affects the initiation of cilia formation. A point mutation was mapped in the L3P line to the gene Rsg1, which encodes a GTPase-like protein. The mutation lies within the GTP-binding pocket and disrupts the highly conserved G1 domain. The mutant protein and other centrosomal and IFT proteins still localize appropriately to the basal body of cilia, suggesting that RSG1 GTPase activity is not required for basal body maturation but is needed for a downstream step in axonemal elongation.
Keywords: RSG1 or CPLANE2; cilia; gene mapping; mouse; neural tube defects.
© 2024 Wiley Periodicals LLC.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

