Serine protease Rv2569c facilitates transmission of Mycobacterium tuberculosis via disrupting the epithelial barrier by cleaving E-cadherin
- PMID: 38722857
- PMCID: PMC11081392
- DOI: 10.1371/journal.ppat.1012214
Serine protease Rv2569c facilitates transmission of Mycobacterium tuberculosis via disrupting the epithelial barrier by cleaving E-cadherin
Abstract
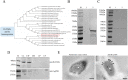
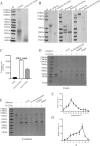
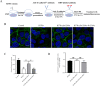
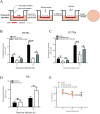
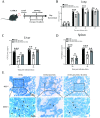
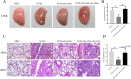
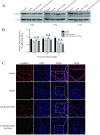
Epithelial cells function as the primary line of defense against invading pathogens. However, bacterial pathogens possess the ability to compromise this barrier and facilitate the transmigration of bacteria. Nonetheless, the specific molecular mechanism employed by Mycobacterium tuberculosis (M.tb) in this process is not fully understood. Here, we investigated the role of Rv2569c in M.tb translocation by assessing its ability to cleave E-cadherin, a crucial component of cell-cell adhesion junctions that are disrupted during bacterial invasion. By utilizing recombinant Rv2569c expressed in Escherichia coli and subsequently purified through affinity chromatography, we demonstrated that Rv2569c exhibited cell wall-associated serine protease activity. Furthermore, Rv2569c was capable of degrading a range of protein substrates, including casein, fibrinogen, fibronectin, and E-cadherin. We also determined that the optimal conditions for the protease activity of Rv2569c occurred at a temperature of 37°C and a pH of 9.0, in the presence of MgCl2. To investigate the function of Rv2569c in M.tb, a deletion mutant of Rv2569c and its complemented strains were generated and used to infect A549 cells and mice. The results of the A549-cell infection experiments revealed that Rv2569c had the ability to cleave E-cadherin and facilitate the transmigration of M.tb through polarized A549 epithelial cell layers. Furthermore, in vivo infection assays demonstrated that Rv2569c could disrupt E-cadherin, enhance the colonization of M.tb, and induce pathological damage in the lungs of C57BL/6 mice. Collectively, these results strongly suggest that M.tb employs the serine protease Rv2569c to disrupt epithelial defenses and facilitate its systemic dissemination by crossing the epithelial barrier.
Copyright: © 2024 Zang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Serine protease Rv2569c inhibits inflammatory response and promotes intracellular survival of Mycobacterium tuberculosis by targeting the RhoG-NF-κB-NLRP3 pathway.Int J Biol Macromol. 2025 May;309(Pt 4):143022. doi: 10.1016/j.ijbiomac.2025.143022. Epub 2025 Apr 12. Int J Biol Macromol. 2025. PMID: 40228763
-
Affinity of Mycobacterium tuberculosis strains for M059K microglial cells after migration through A549 alveolar epithelium.Eur J Clin Microbiol Infect Dis. 2021 Sep;40(9):1881-1889. doi: 10.1007/s10096-021-04226-1. Epub 2021 Apr 9. Eur J Clin Microbiol Infect Dis. 2021. PMID: 33834319
-
Cleavage of E-cadherin by porcine respiratory bacterial pathogens facilitates airway epithelial barrier disruption and bacterial paracellular transmigration.Virulence. 2021 Dec;12(1):2296-2313. doi: 10.1080/21505594.2021.1966996. Virulence. 2021. PMID: 34482810 Free PMC article.
-
Mycobacterium tuberculosis Primary Infection and Dissemination: A Critical Role for Alveolar Epithelial Cells.Front Cell Infect Microbiol. 2019 Aug 21;9:299. doi: 10.3389/fcimb.2019.00299. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31497538 Free PMC article. Review.
-
Mycobacterium tuberculosis proteases and implications for new antibiotics against tuberculosis.Crit Rev Eukaryot Gene Expr. 2011;21(4):347-61. doi: 10.1615/critreveukargeneexpr.v21.i4.50. Crit Rev Eukaryot Gene Expr. 2011. PMID: 22181704 Review.
Cited by
-
Role of E-cadherin in epithelial barrier dysfunction: implications for bacterial infection, inflammation, and disease pathogenesis.Front Cell Infect Microbiol. 2025 Feb 11;15:1506636. doi: 10.3389/fcimb.2025.1506636. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40007608 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

