Integrative multi-omics analyses to identify the genetic and functional mechanisms underlying ovarian cancer risk regions
- PMID: 38723632
- PMCID: PMC11179261
- DOI: 10.1016/j.ajhg.2024.04.011
Integrative multi-omics analyses to identify the genetic and functional mechanisms underlying ovarian cancer risk regions
Abstract
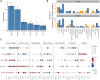
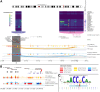
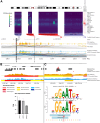
To identify credible causal risk variants (CCVs) associated with different histotypes of epithelial ovarian cancer (EOC), we performed genome-wide association analysis for 470,825 genotyped and 10,163,797 imputed SNPs in 25,981 EOC cases and 105,724 controls of European origin. We identified five histotype-specific EOC risk regions (p value <5 × 10-8) and confirmed previously reported associations for 27 risk regions. Conditional analyses identified an additional 11 signals independent of the primary signal at six risk regions (p value <10-5). Fine mapping identified 4,008 CCVs in these regions, of which 1,452 CCVs were located in ovarian cancer-related chromatin marks with significant enrichment in active enhancers, active promoters, and active regions for CCVs from each EOC histotype. Transcriptome-wide association and colocalization analyses across histotypes using tissue-specific and cross-tissue datasets identified 86 candidate susceptibility genes in known EOC risk regions and 32 genes in 23 additional genomic regions that may represent novel EOC risk loci (false discovery rate <0.05). Finally, by integrating genome-wide HiChIP interactome analysis with transcriptome-wide association study (TWAS), variant effect predictor, transcription factor ChIP-seq, and motifbreakR data, we identified candidate gene-CCV interactions at each locus. This included risk loci where TWAS identified one or more candidate susceptibility genes (e.g., HOXD-AS2, HOXD8, and HOXD3 at 2q31) and other loci where no candidate gene was identified (e.g., MYC and PVT1 at 8q24) by TWAS. In summary, this study describes a functional framework and provides a greater understanding of the biological significance of risk alleles and candidate gene targets at EOC susceptibility loci identified by a genome-wide association study.
Keywords: GWAS; epithelial ovarian cancer risk; fine mapping; functional mechanisms.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
Cis- and trans-eQTL TWASs of breast and ovarian cancer identify more than 100 susceptibility genes in the BCAC and OCAC consortia.Am J Hum Genet. 2024 Jun 6;111(6):1084-1099. doi: 10.1016/j.ajhg.2024.04.012. Epub 2024 May 8. Am J Hum Genet. 2024. PMID: 38723630 Free PMC article.
References
-
- Song H., Cicek M.S., Dicks E., Harrington P., Ramus S.J., Cunningham J.M., Fridley B.L., Tyrer J.P., Alsop J., Jimenez-Linan M., et al. The contribution of deleterious germline mutations in BRCA1, BRCA2 and the mismatch repair genes to ovarian cancer in the population. Hum. Mol. Genet. 2014;23:4703–4709. - PMC - PubMed
-
- Vang R., Shih I.-M., Kurman R.J. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology. 2013;62:44–58. - PubMed
-
- Gounaris I., Brenton J.D. Molecular pathogenesis of ovarian clear cell carcinoma. Future Oncol. 2015;11:1389–1405. - PubMed

