Landscape of the gut mycobiome dynamics during pregnancy and its relationship with host metabolism and pregnancy health
- PMID: 38724219
- PMCID: PMC11287620
- DOI: 10.1136/gutjnl-2024-332260
Landscape of the gut mycobiome dynamics during pregnancy and its relationship with host metabolism and pregnancy health
Abstract
Objective: The remodelling of gut mycobiome (ie, fungi) during pregnancy and its potential influence on host metabolism and pregnancy health remains largely unexplored. Here, we aim to examine the characteristics of gut fungi in pregnant women, and reveal the associations between gut mycobiome, host metabolome and pregnancy health.
Design: Based on a prospective birth cohort in central China (2017 to 2020): Tongji-Huaxi-Shuangliu Birth Cohort, we included 4800 participants who had available ITS2 sequencing data, dietary information and clinical records during their pregnancy. Additionally, we established a subcohort of 1059 participants, which included 514 women who gave birth to preterm, low birthweight or macrosomia infants, as well as 545 randomly selected controls. In this subcohort, a total of 750, 748 and 709 participants had ITS2 sequencing data, 16S sequencing data and serum metabolome data available, respectively, across all trimesters.
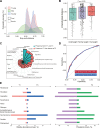
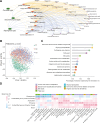
Results: The composition of gut fungi changes dramatically from early to late pregnancy, exhibiting a greater degree of variability and individuality compared with changes observed in gut bacteria. The multiomics data provide a landscape of the networks among gut mycobiome, biological functionality, serum metabolites and pregnancy health, pinpointing the link between Mucor and adverse pregnancy outcomes. The prepregnancy overweight status is a key factor influencing both gut mycobiome compositional alteration and the pattern of metabolic remodelling during pregnancy.
Conclusion: This study provides a landscape of gut mycobiome dynamics during pregnancy and its relationship with host metabolism and pregnancy health, which lays the foundation of the future gut mycobiome investigation for healthy pregnancy.
Keywords: INTESTINAL MICROBIOLOGY.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
