A common polymorphism in the Intelectin-1 gene influences mucus plugging in severe asthma
- PMID: 38724552
- PMCID: PMC11082194
- DOI: 10.1038/s41467-024-48034-5
A common polymorphism in the Intelectin-1 gene influences mucus plugging in severe asthma
Abstract
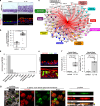
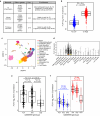
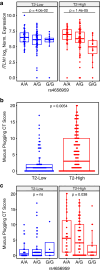
By incompletely understood mechanisms, type 2 (T2) inflammation present in the airways of severe asthmatics drives the formation of pathologic mucus which leads to airway mucus plugging. Here we investigate the molecular role and clinical significance of intelectin-1 (ITLN-1) in the development of pathologic airway mucus in asthma. Through analyses of human airway epithelial cells we find that ITLN1 gene expression is highly induced by interleukin-13 (IL-13) in a subset of metaplastic MUC5AC+ mucus secretory cells, and that ITLN-1 protein is a secreted component of IL-13-induced mucus. Additionally, we find ITLN-1 protein binds the C-terminus of the MUC5AC mucin and that its deletion in airway epithelial cells partially reverses IL-13-induced mucostasis. Through analysis of nasal airway epithelial brushings, we find that ITLN1 is highly expressed in T2-high asthmatics, when compared to T2-low children. Furthermore, we demonstrate that both ITLN-1 gene expression and protein levels are significantly reduced by a common genetic variant that is associated with protection from the formation of mucus plugs in T2-high asthma. This work identifies an important biomarker and targetable pathways for the treatment of mucus obstruction in asthma.
© 2024. The Author(s).
Conflict of interest statement
On behalf of the SARP study: The following companies provided financial support for study activities at the Coordinating and Clinical Centers beyond the third year of SARP patient follow-up: AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi–Genzyme–Regeneron, and TEVA. These companies had no role in study design, data collection, or data analysis, nor preparation of the manuscript or decision to publish, and the only restriction on the funds was that they be used to support the SARP initiative. The authors of this manuscript report the following COIs: J.L.E. is listed as an inventor on the filed patent application by National Jewish Health, PCT/US24/11470 titled “Methods and Compositions for Treating Mucus Obstruction in Severe Asthma.” V.E.O. has received compensation for his role in Independent Data and Monitoring Committees for trials sponsored by Regeneron and Sanofi. B.M.E. receives consulting fees from Aer Therapeutics LLC, and receives stocks for service on the company board. B.M.E. is listed as one of the inventors on a filed patent application for lung image-based scoring systems that quantify mucus plug burden in the lung. T.S.H. is a stockholder and advisor for Aer Therapeutics LLC. M. Castro reports grants/research support from NIH, ALA, PCORI, AstraZeneca, Gala Therapeutics, Genentech, GSK, Novartis, Pulmatrix, Sanofi-Aventis, Shionogi, and Theravance. M. Castro receives consulting fees from Allakos, Amgen, Arrowhead Pharmaceuticals, Genentech, GSK, Merck, Novartis, OM Pharma, Pfizer, Pioneering Medicines, Sanofi-Aventis, and Teva, and receives payment for speaker’s bureau activities from Amgen, AstraZeneca, Genentech, Regeneron, Sanofi-Aventis, and Teva. M. Castro receives stock options from Aer Therapeutics. E.I. reports grants/research support or research materials from NHLBI, NIH, AstraZeneca, Avillion, Gossamer Bio, PCORI, Circassia, Genentech, TEVA, Sun Pharma, Laurel Pharmaceuticals, Om Pharma, Nestle, CSL Behring, AMSA, and Sanofi-Regeneron. E.I. receives consulting fees from AB Science, Allergy and Asthma Network, Amgen, Arrowhead Pharmaceuticals, AstraZeneca, GlaxoSmithKline, Merck, NHLBI, PPS Health, Regeneron, Sanofi Genzyme, TEVA, Cowen, Guidepoint, Windrose Consulting Group, and Reach Market Research. E.I. reports royalties from Wolters Kluwer, and speaker fees from Westchester Medical Center and Clearview Health Partners. E.I. has provided expert testimony for Cambridge Medical Experts. E.I. has received compensation for participation in the Data Safety Monitoring Board for Novartis, as a member of the coordinating committee in NAEPP: National Asthma Education Prevention Program, and stock options for Vorso. B.D.L., D.T.M., K.S., and L.D. reports grant funding support for SARP study activities at the Coordinating and Clinical Centers beyond the third year of SARP patient follow-up from AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi–Genzyme–Regeneron, and TEVA. K.S. has received advisory fees from AstraZeneca. D.S.G. is listed as one of the inventors on a filed patent application for lung image-based scoring systems that quantify mucus plug burden in the lung. A.T.H. has received grants/research support for this study from NHLBI, and for SARP study activities at the Coordinating and Clinical Centers beyond the third year of SARP patient follow-up from AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi–Genzyme–Regeneron, and TEVA. W.C.M. reports grant funding support from NHLI and ALA, and reports grant funding support for SARP study activities at the Coordinating and Clinical Centers beyond the third year of SARP patient follow-up from AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi–Genzyme–Regeneron, and TEVA. L.D. also reports grant funding from NHLBI and ALA-ACRC, consulting fees from OM Pharma, research support/research equipment and materials from GlaxoSmithKline, Laurel, Sun Pharma, Vifor/OM Pharma, Vitaeris/CSL Behring, and Vitaflo. N.N.J. has previously received consulting fees from GlaxoSmithKline and AstraZeneca. S.E.W. is a Primary Investigator on an ongoing investigational research study with Regeneron. P.G.W. is a consultant with Regeneron, Sanofi, AstraZeneca, and Roche. J.V.F. reports grant funding from NIH/NHLBI, and reports grant funding support for SARP study activities at the Coordinating and Clinical Centers beyond the third year of SARP patient follow-up from AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi–Genzyme–Regeneron, and TEVA. J.V.F. is the inventor of granted patents describing thiol modified saccharides as drugs to treat mucus-associated lung disease, including asthma, patent numbers US 9,856,283; US 910,526,359; US 11,021,506. J.V.F. is listed as one of the inventors on a filed patent application for lung image-based scoring systems that quantify mucus plug burden in the lung. J.V.F. is a member of the Scientific Advisory Board and receives financial and stock options as compensation for services on the Advisory Board for Suzhou Connect Biopharmaceuticals, Ltd. J.V.F. is a member of the company board and is a consultant for Aer Therapeutics, and receives financial compensation and stocks provided as compensation for services provided. M.A.S. has received speaker fees from the Colorado Allergy and Asthma Society, and is a consultant for Escient Pharmaceuticals. M.A.S. is listed as an inventor on the filed patent application by National Jewish Health, PCT/US24/11470 titled “Methods and Compositions for Treating Mucus Obstruction in Severe Asthma.” S.P.S., M.A.L., N.D.J., E.H.C., M.C.P., M. Chioccioli, C.M.M., B.B.P., N.D., R.P., C.R., M.T.M., C.E., J.R.E., A.C.Y.M., D.H., S.H., S.S., L.F., A.F.M., G.L.Z., C.R.M., J.G., X.Z., V.M., H.W.C., P.C., E.D.G., P.Z., N.R., E.M.D., M.T., E.R.B., S.C.E., D.A.M., M.L.S., J.R.S., C.G.P., and E.G.B. declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL107202/HL/NHLBI NIH HHS/United States
- U01 HL146002/HL/NHLBI NIH HHS/United States
- K24 HL137013/HL/NHLBI NIH HHS/United States
- R01 HL135156/HL/NHLBI NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- R01 HL080414/HL/NHLBI NIH HHS/United States
- U10 HL109146/HL/NHLBI NIH HHS/United States
- R01 HL128439/HL/NHLBI NIH HHS/United States
- S10 OD028538/OD/NIH HHS/United States
- P01 HL132821/HL/NHLBI NIH HHS/United States
- R01 MD010443/MD/NIMHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

