Supramolecular Nanofibers Ameliorate Bleomycin-Induced Pulmonary Fibrosis by Restoring Autophagy
- PMID: 38725147
- PMCID: PMC11267363
- DOI: 10.1002/advs.202401327
Supramolecular Nanofibers Ameliorate Bleomycin-Induced Pulmonary Fibrosis by Restoring Autophagy
Abstract
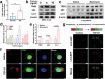
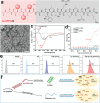
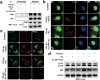
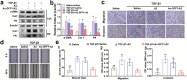
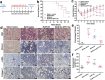
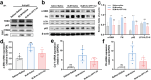
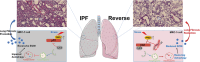
Idiopathic pulmonary fibrosis (IPF) is a progressive and ultimately fatal interstitial lung disease, with limited therapeutic options available. Impaired autophagy resulting from aberrant TRB3/p62 protein-protein interactions (PPIs) contributes to the progression of IPF. Restoration of autophagy by modulating the TRB3/p62 PPIs has rarely been reported for the treatment of IPF. Herein, peptide nanofibers are developed that specifically bind to TRB3 protein and explored their potential as a therapeutic approach for IPF. By conjugating with the self-assembling fragment (Ac-GFFY), a TRB3-binding peptide motif A2 allows for the formation of nanofibers with a stable α-helix secondary structure. The resulting peptide (Ac-GFFY-A2) nanofibers exhibit specific high-affinity binding to TRB3 protein in saline buffer and better capacity of cellular uptake to A2 peptide. Furthermore, the TRB3-targeting peptide nanofibers efficiently interfere with the aberrant TRB3/p62 PPIs in activated fibroblasts and fibrotic lung tissue of mice, thereby restoring autophagy dysfunction. The TRB3-targeting peptide nanofibers inhibit myofibroblast differentiation, collagen production, and fibroblast migration in vitro is demonstrated, as well as bleomycin-induced pulmonary fibrosis in vivo. This study provides a supramolecular method to modulate PPIs and highlights a promising strategy for treating IPF diseases by restoring autophagy.
Keywords: autophagy; peptide nanofiber; protein‐protein interactions; pulmonary fibrosis; self‐assembly.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
