A likelihood ratio approach for utilizing case-control data in the clinical classification of rare sequence variants: application to BRCA1 and BRCA2
- PMID: 38725546
- PMCID: PMC11080979
- DOI: 10.1155/2023/9961341
A likelihood ratio approach for utilizing case-control data in the clinical classification of rare sequence variants: application to BRCA1 and BRCA2
Abstract
A large number of variants identified through clinical genetic testing in disease susceptibility genes, are of uncertain significance (VUS). Following the recommendations of the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP), the frequency in case-control datasets (PS4 criterion), can inform their interpretation. We present a novel case-control likelihood ratio-based method that incorporates gene-specific age-related penetrance. We demonstrate the utility of this method in the analysis of simulated and real datasets. In the analyses of simulated data, the likelihood ratio method was more powerful compared to other methods. Likelihood ratios were calculated for a case-control dataset of BRCA1 and BRCA2 variants from the Breast Cancer Association Consortium (BCAC), and compared with logistic regression results. A larger number of variants reached evidence in favor of pathogenicity, and a substantial number of variants had evidence against pathogenicity - findings that would not have been reached using other case-control analysis methods. Our novel method provides greater power to classify rare variants compared to classical case-control methods. As an initiative from the ENIGMA Analytical Working Group, we provide user-friendly scripts and pre-formatted excel calculators for implementation of the method for rare variants in BRCA1, BRCA2 and other high-risk genes with known penetrance.
Keywords: ACMG/AMP; BRCA; PS4; VUS; case-control; likelihood ratio; variant classification.
Conflict of interest statement
The following authors declare conflicts not directly relevant to this work as stated below. Usha Menon has a patent (no: EP10178345.4) for Breast Cancer Diagnostics and held personal shares in Abcodia between 1st April 2011 and 30 October 2021. She is a member of the Research Advisory Panel, Yorkshire Cancer Research, Trial Steering Committee, NOVEL, and Scientific Advisory Board of Tina's Wish. She has received grants from the Medical Research Council (MRC), Cancer Research UK, the National Institute for Health Research (NIHR), and The Eve Appeal. She is part of research collaborations with iLOF, RNG Guardian and Micronoma. All other authors declare that they have no conflict of interests.
Figures
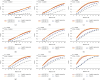
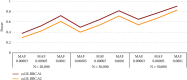
References
-
- Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine . 2015;17(5):405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- Goldgar D. E., Easton D. F., Deffenbaugh A. M., Monteiro A. N., Tavtigian S. V., Couch F. J. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. The American Journal of Human Genetics . 2004;75(4):535–544. doi: 10.1086/424388. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA176785/CA/NCI NIH HHS/United States
- NU58DP006344/DP/NCCDPHP CDC HHS/United States
- U01 CA176726/CA/NCI NIH HHS/United States
- R01 CA058860/CA/NCI NIH HHS/United States
- G1000143/MRC_/Medical Research Council/United Kingdom
- P50 CA116201/CA/NCI NIH HHS/United States
- HHSN261201800015C/CA/NCI NIH HHS/United States
- R01 CA192393/CA/NCI NIH HHS/United States
- HHSN261201800009I/CA/NCI NIH HHS/United States
- MR/M012190/1/MRC_/Medical Research Council/United Kingdom
- UM1 CA164917/CA/NCI NIH HHS/United States
- U01 CA199277/CA/NCI NIH HHS/United States
- U01 CA179715/CA/NCI NIH HHS/United States
- R01 CA128931/CA/NCI NIH HHS/United States
- HHSN261201800032C/CA/NCI NIH HHS/United States
- U54 CA156733/CA/NCI NIH HHS/United States
- HHSN261201800009C/CA/NCI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- UM1 CA164973/CA/NCI NIH HHS/United States
- Z01 ES044005/ImNIH/Intramural NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- UM1 CA164920/CA/NCI NIH HHS/United States
- R01 CA097396/CA/NCI NIH HHS/United States
- UM1 CA176726/CA/NCI NIH HHS/United States
- Z01 ES049033/ImNIH/Intramural NIH HHS/United States
- HHSN261201800015I/CA/NCI NIH HHS/United States
- Z01 ES049030/ImNIH/Intramural NIH HHS/United States
- K07 CA092044/CA/NCI NIH HHS/United States
- P50 CA058223/CA/NCI NIH HHS/United States
- R01 CA100374/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 CA128978/CA/NCI NIH HHS/United States
- U19 CA148537/CA/NCI NIH HHS/United States
- R01 CA116167/CA/NCI NIH HHS/United States
- R01 CA177150/CA/NCI NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- UM1 CA186107/CA/NCI NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- R01 CA077398/CA/NCI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- R01 CA132839/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- U01 CA058860/CA/NCI NIH HHS/United States
- U01 CA164920/CA/NCI NIH HHS/United States
- Z01 CP010119/ImNIH/Intramural NIH HHS/United States
- R35 CA253187/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- HHSN261201800032I/CA/NCI NIH HHS/United States
- U01 CA098758/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- U01 CA164973/CA/NCI NIH HHS/United States
- R01 CA140286/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

