Immune Cells Are Differentially Affected by SARS-CoV-2 Viral Loads in K18-hACE2 Mice
- PMID: 38725670
- PMCID: PMC11076298
- DOI: 10.4110/in.2024.24.e7
Immune Cells Are Differentially Affected by SARS-CoV-2 Viral Loads in K18-hACE2 Mice
Abstract
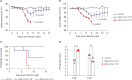
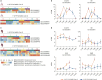
Viral load and the duration of viral shedding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are important determinants of the transmission of coronavirus disease 2019. In this study, we examined the effects of viral doses on the lung and spleen of K18-hACE2 transgenic mice by temporal histological and transcriptional analyses. Approximately, 1×105 plaque-forming units (PFU) of SARS-CoV-2 induced strong host responses in the lungs from 2 days post inoculation (dpi) which did not recover until the mice died, whereas responses to the virus were obvious at 5 days, recovering to the basal state by 14 dpi at 1×102 PFU. Further, flow cytometry showed that number of CD8+ T cells continuously increased in 1×102 PFU-virus-infected lungs from 2 dpi, but not in 1×105 PFU-virus-infected lungs. In spleens, responses to the virus were prominent from 2 dpi, and number of B cells was significantly decreased at 1×105 PFU; however, 1×102 PFU of virus induced very weak responses from 2 dpi which recovered by 10 dpi. Although the defense responses returned to normal and the mice survived, lung histology showed evidence of fibrosis, suggesting sequelae of SARS-CoV-2 infection. Our findings indicate that specific effectors of the immune response in the lung and spleen were either increased or depleted in response to doses of SARS-CoV-2. This study demonstrated that the response of local and systemic immune effectors to a viral infection varies with viral dose, which either exacerbates the severity of the infection or accelerates its elimination.
Keywords: Dose-response relationship, immunologic; Immune response; K18-hACE2 mice; SARS-CoV-2; Transcriptome profiling.
Copyright © 2024. The Korean Association of Immunologists.
Conflict of interest statement
Conflict of Interest: The authors declare no potential conflicts of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

