Genome-wide mapping of G-quadruplex DNA: a step-by-step guide to select the most effective method
- PMID: 38725910
- PMCID: PMC11078208
- DOI: 10.1039/d4cb00023d
Genome-wide mapping of G-quadruplex DNA: a step-by-step guide to select the most effective method
Abstract
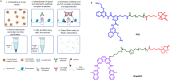
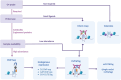
The development of methods that enabled genome-wide mapping of DNA G-quadruplex structures in chromatin has played a critical role in providing evidence to support the formation of these structures in living cells. Over the past decade, a variety of methods aimed at mapping G-quadruplexes have been reported in the literature. In this critical review, we have sought to provide a technical overview on the relative strengths and weaknesses of the genomics approaches currently available, offering step-by-step guidance to assessing experimental needs and selecting the most appropriate method to achieve effective genome-wide mapping of DNA G-quadruplexes.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures









References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

