Proteomics of human platelet lysates and insight from animal studies on platelet protein diffusion to hippocampus upon intranasal administration
- PMID: 38726021
- PMCID: PMC11080963
- DOI: 10.1063/5.0196553
Proteomics of human platelet lysates and insight from animal studies on platelet protein diffusion to hippocampus upon intranasal administration
Abstract
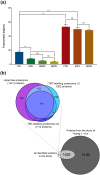
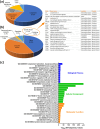
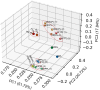
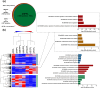
Human platelet lysates (HPLs) from allogeneic platelet concentrates (PCs) are biomaterials, which are rich in various trophic factors, increasingly used in regenerative medicine and biotherapy. Understanding how preparation methods influence the HPL protein profile, biological function, and clinical outcomes is crucial. Our study sheds light on the proteomes and functionality of different HPLs, with the aim of advancing their scientifically grounded clinical applications. To achieve this, PCs suspended in plasma underwent three distinct processing methods, resulting in seven HPL types. We used three characterization techniques: label-free proteomics and tandem mass tag (TMT)-based quantitative proteomics, both before and after the immunodepletion of abundant plasma proteins. Bioinformatic tools assessed the proteome, and western blotting validated our quantitative proteomics data. Subsequent pre-clinical studies with fluorescent labeling and label-free proteomics were used as a proof of concept for brain diffusion. Our findings revealed 1441 proteins detected using the label-free method, 952 proteins from the TMT experiment before and after depletion, and 1114 proteins from the subsequent TMT experiment on depleted HPLs. Most detected proteins were cytoplasmic, playing key roles in catalysis, hemostasis, and immune responses. Notably, the processing methodologies significantly influenced HPL compositions, their canonical pathways, and, consequently, their functionality. Each HPL exhibited specific abundant proteins, providing valuable insight for tailored clinical applications. Immunoblotting results for selected proteins corroborated our quantitative proteomics data. The diffusion and differential effects to the hippocampus of a neuroprotective HPL administered intranasally to mice were demonstrated. This proteomics study advances our understanding of HPLs, suggesting ways to standardize and customize their production for better clinical efficacy in regenerative medicine and biotherapy. Proteomic analyses also offered objective evidence that HPPL, upon intranasal delivery, not only effectively diffuses to the hippocampus but also alters protein expression in mice, bolstering its potential as a treatment for memory impairments.
© 2024 Author(s).
Conflict of interest statement
T.B. is one co-founder of Invenis Biotherapies and is listed as the inventor on patent applications owned by Taipei Medical University and University of Lille. The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Influence of platelet storage time on human platelet lysates and platelet lysate-expanded mesenchymal stromal cells for bone tissue engineering.Stem Cell Res Ther. 2020 Sep 23;11(1):351. doi: 10.1186/s13287-020-01863-9. Stem Cell Res Ther. 2020. PMID: 32962723 Free PMC article.
-
Expanding applications of allogeneic platelets, platelet lysates, and platelet extracellular vesicles in cell therapy, regenerative medicine, and targeted drug delivery.J Biomed Sci. 2023 Sep 14;30(1):79. doi: 10.1186/s12929-023-00972-w. J Biomed Sci. 2023. PMID: 37704991 Free PMC article. Review.
-
Neuroprotective activity of a virus-safe nanofiltered human platelet lysate depleted of extracellular vesicles in Parkinson's disease and traumatic brain injury models.Bioeng Transl Med. 2022 Jul 12;8(1):e10360. doi: 10.1002/btm2.10360. eCollection 2023 Jan. Bioeng Transl Med. 2022. PMID: 36684076 Free PMC article.
-
Characterization and Chromatographic Isolation of Platelet Extracellular Vesicles from Human Platelet Lysates for Applications in Neuroregenerative Medicine.ACS Biomater Sci Eng. 2021 Dec 13;7(12):5823-5835. doi: 10.1021/acsbiomaterials.1c01226. Epub 2021 Nov 30. ACS Biomater Sci Eng. 2021. PMID: 34846835
-
Proteome changes in platelets after pathogen inactivation--an interlaboratory consensus.Transfus Med Rev. 2014 Apr;28(2):72-83. doi: 10.1016/j.tmrv.2014.02.002. Epub 2014 Feb 25. Transfus Med Rev. 2014. PMID: 24685438 Review.
Cited by
-
Bioprocessing of human platelet concentrates to generate lysates and extracellular vesicles for therapeutic applications.MethodsX. 2024 Jul 9;13:102822. doi: 10.1016/j.mex.2024.102822. eCollection 2024 Dec. MethodsX. 2024. PMID: 39105089 Free PMC article.
-
Pilot study characterizing a single pooled preparation of equine platelet lysate for nebulization in the horse.Front Vet Sci. 2024 Dec 12;11:1488942. doi: 10.3389/fvets.2024.1488942. eCollection 2024. Front Vet Sci. 2024. PMID: 39726585 Free PMC article.
-
Neuroprotective effects of intranasal extracellular vesicles from human platelet concentrates supernatants in traumatic brain injury and Parkinson's disease models.J Biomed Sci. 2024 Sep 5;31(1):87. doi: 10.1186/s12929-024-01072-z. J Biomed Sci. 2024. PMID: 39237980 Free PMC article.
-
Human platelet lysate: a potential therapeutic for intracerebral hemorrhage.Front Neurosci. 2025 Jan 15;18:1517601. doi: 10.3389/fnins.2024.1517601. eCollection 2024. Front Neurosci. 2025. PMID: 39881806 Free PMC article. Review.
References
-
- Burnouf T., Chou M. L., Lundy D. J., Chuang E. Y., Tseng C. L., and Goubran H., “ Expanding applications of allogeneic platelets, platelet lysates, and platelet extracellular vesicles in cell therapy, regenerative medicine, and targeted drug delivery,” J. Biomed. Sci. 30, 79 (2023).10.1186/s12929-023-00972-w - DOI - PMC - PubMed
-
- Schallmoser K., Henschler R., Gabriel C., Koh M. B. C., and Burnouf T., “ Production and quality requirements of human platelet lysate: A position statement from the working party on cellular therapies of the international society of blood transfusion,” Trends Biotechnol. 38, 13–23 (2020).10.1016/j.tibtech.2019.06.002 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

