Transcription factor STAT4 counteracts radiotherapy resistance in breast carcinoma cells by activating the MALAT1/miR-21-5p/THRB regulatory network
- PMID: 38726265
- PMCID: PMC11076251
- DOI: 10.62347/VSJU7227
Transcription factor STAT4 counteracts radiotherapy resistance in breast carcinoma cells by activating the MALAT1/miR-21-5p/THRB regulatory network
Abstract
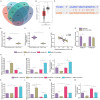
Considering the limited research and the prevailing evidence of STAT4's tumor-suppressing role in breast carcinoma (BC) or in breast radiotherapy (RT) sensitivity requires more in-depth exploration. Our study delves into how STAT4, a transcription factor, affects BC cell resistance to radiotherapy by regulating the MALAT1/miR-21-5p/THRB axis. Bioinformatics analysis was performed to predict the regulatory mechanisms associated with STAT4 in BC. Subsequently, we identified the expression profiles of STAT4, MALAT1, miR-21-5p, and THRB in various tissues and cell lines, exploring their interactions and impact on RT resistance in BC cells. Moreover, animal models were established with X-ray irradiation for further validation. We discovered that STAT4, which is found to be minimally expressed in breast carcinoma (BC) tissues and cell lines, has been associated with a poorer prognosis. In vitro cellular assays indicated that STAT4 could mitigate radiotherapy resistance in BC cells by transcriptional activation of MALAT1. Additionally, MALAT1 up-regulated THRB expression by adsorbing miR-21-5p. As demonstrated in vitro and in vivo, overexpressing STAT4 inhibited miR-21-5p and enhanced THRB levels through transcriptional activation of MALAT1, which ultimately contributes to the reversal of radiotherapy resistance in BC cells and the suppression of tumor formation in nude mice. Collectively, STAT4 could inhibit miR-21-5p and up-regulate THRB expression through transcriptional activation of MALAT1, thereby mitigating BC cell resistance to radiotherapy and ultimately preventing BC development and progression.
Keywords: Breast carcinoma; MALAT1; STAT4; THRB; long non-coding RNA; microRNA-21-5p; radiotherapy resistance; transcription factor.
AJCR Copyright © 2024.
Conflict of interest statement
None.
Figures








References
-
- Aghiorghiesei O, Zanoaga O, Raduly L, Aghiorghiesei AI, Chiroi P, Trif A, Buiga R, Budisan L, Lucaciu O, Pop LA, Braicu C, Campian R, Berindan-Neagoe I. Dysregulation of miR-21-5p, miR-93-5p, miR-200c-3p and miR-205-5p in oral squamous cell carcinoma: a potential biomarkers panel? Curr Issues Mol Biol. 2022;44:1754–1767. - PMC - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. - PubMed
-
- Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E, Farahmand L. Breast cancer: biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
