Small-Molecule Inhibitors of TIPE3 Protein Identified through Deep Learning Suppress Cancer Cell Growth In Vitro
- PMID: 38727307
- PMCID: PMC11082981
- DOI: 10.3390/cells13090771
Small-Molecule Inhibitors of TIPE3 Protein Identified through Deep Learning Suppress Cancer Cell Growth In Vitro
Abstract
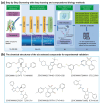
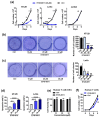
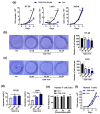
Tumor necrosis factor-α-induced protein 8-like 3 (TNFAIP8L3 or TIPE3) functions as a transfer protein for lipid second messengers. TIPE3 is highly upregulated in several human cancers and has been established to significantly promote tumor cell proliferation, migration, and invasion and inhibit the apoptosis of cancer cells. Thus, inhibiting the function of TIPE3 is expected to be an effective strategy against cancer. The advancement of artificial intelligence (AI)-driven drug development has recently invigorated research in anti-cancer drug development. In this work, we incorporated DFCNN, Autodock Vina docking, DeepBindBC, MD, and metadynamics to efficiently identify inhibitors of TIPE3 from a ZINC compound dataset. Six potential candidates were selected for further experimental study to validate their anti-tumor activity. Among these, three small-molecule compounds (K784-8160, E745-0011, and 7238-1516) showed significant anti-tumor activity in vitro, leading to reduced tumor cell viability, proliferation, and migration and enhanced apoptotic tumor cell death. Notably, E745-0011 and 7238-1516 exhibited selective cytotoxicity toward tumor cells with high TIPE3 expression while having little or no effect on normal human cells or tumor cells with low TIPE3 expression. A molecular docking analysis further supported their interactions with TIPE3, highlighting hydrophobic interactions and their shared interaction residues and offering insights for designing more effective inhibitors. Taken together, this work demonstrates the feasibility of incorporating deep learning and MD simulations in virtual drug screening and provides inhibitors with significant potential for anti-cancer drug development against TIPE3-.
Keywords: TIPE3; anti-tumor; deep learning; small-molecule compounds.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Padmavathi G., Banik K., Monisha J., Bordoloi D., Shabnam B., Arfuso F., Sethi G., Fan L., Kunnumakkara A.B. Novel tumor necrosis factor-α induced protein eight (TNFAIP8/TIPE) family: Functions and downstream targets involved in cancer progression. Cancer Lett. 2018;432:260–271. doi: 10.1016/j.canlet.2018.06.017. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2022YFA0912400/the National Key R&D Program of China
- 32130040/the National Natural Science Foundation of China
- 62106253/the National Natural Science Foundation of China
- 21933010/the National Natural Science Foundation of China
- 22250710136/the National Natural Science Foundation of China
- 22333006/the National Natural Science Foundation of China
- 82250710172/the National Natural Science Foundation of China
- 2021A1515110053/the Guangdong Basic and Applied Basic Research Foundation
- JCYJ20220818100806015/the Shenzhen Basic Science Research Project
- JCYJ20220818100804009/the Shenzhen Basic Science Research Project
- JCYJ20210324101402007/the Shenzhen Basic Science Research Project
- B2301006/the Shenzhen Medical Research Fund
LinkOut - more resources
Full Text Sources

