A polygenic risk score for alcohol-associated cirrhosis among heavy drinkers with European ancestry
- PMID: 38727677
- PMCID: PMC11093576
- DOI: 10.1097/HC9.0000000000000431
A polygenic risk score for alcohol-associated cirrhosis among heavy drinkers with European ancestry
Abstract
Background: Polygenic Risk Scores (PRS) based on results from genome-wide association studies offer the prospect of risk stratification for many common and complex diseases. We developed a PRS for alcohol-associated cirrhosis by comparing single-nucleotide polymorphisms among patients with alcohol-associated cirrhosis (ALC) versus drinkers who did not have evidence of liver fibrosis/cirrhosis.
Methods: Using a data-driven approach, a PRS for ALC was generated using a meta-genome-wide association study of ALC (N=4305) and an independent cohort of heavy drinkers with ALC and without significant liver disease (N=3037). It was validated in 2 additional independent cohorts from the UK Biobank with diagnosed ALC (N=467) and high-risk drinking controls (N=8981) and participants in the Indiana Biobank Liver cohort with alcohol-associated liver disease (N=121) and controls without liver disease (N=3239).
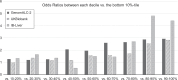
Results: A 20-single-nucleotide polymorphisms PRS for ALC (PRSALC) was generated that stratified risk for ALC comparing the top and bottom deciles of PRS in the 2 validation cohorts (ORs: 2.83 [95% CI: 1.82 -4.39] in UK Biobank; 4.40 [1.56 -12.44] in Indiana Biobank Liver cohort). Furthermore, PRSALC improved the prediction of ALC risk when added to the models of clinically known predictors of ALC risk. It also stratified the risk for metabolic dysfunction -associated steatotic liver disease -cirrhosis (3.94 [2.23 -6.95]) in the Indiana Biobank Liver cohort -based exploratory analysis.
Conclusions: PRSALC incorporates 20 single-nucleotide polymorphisms, predicts increased risk for ALC, and improves risk stratification for ALC compared with the models that only include clinical risk factors. This new score has the potential for early detection of heavy drinking patients who are at high risk for ALC.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Naga P. Chalasani has a number of consulting agreements with and research grants from the pharmaceutical industry, but they are not significantly or directly related to this paper. Munir Pirmohamed receives research funding from various organizations, including the MRC and NIHR. He has also received partnership funding for the following: MRC Clinical Pharmacology Training Scheme (co-funded by MRC and Roche, UCB, Eli Lilly and Novartis); a PhD studentship jointly funded by EPSRC and Astra Zeneca; and grant funding from Vistagen Therapeutics. He has also unrestricted educational grant support for the UK Pharmacogenetics and Stratified Medicine Network from Bristol-Myers Squibb and UCB. He has developed an HLA genotyping panel with MC Diagnostics but does not benefit financially from this. He is part of the IMI Consortium ARDAT (
Figures