Binucleated human hepatocytes arise through late cytokinetic regression during endomitosis M phase
- PMID: 38727809
- PMCID: PMC11090133
- DOI: 10.1083/jcb.202403020
Binucleated human hepatocytes arise through late cytokinetic regression during endomitosis M phase
Abstract
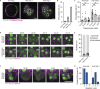
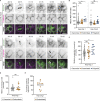
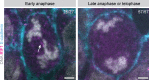
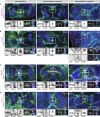
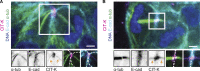
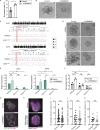
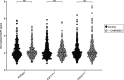
Binucleated polyploid cells are common in many animal tissues, where they arise by endomitosis, a non-canonical cell cycle in which cells enter M phase but do not undergo cytokinesis. Different steps of cytokinesis have been shown to be inhibited during endomitosis M phase in rodents, but it is currently unknown how human cells undergo endomitosis. In this study, we use fetal-derived human hepatocyte organoids (Hep-Orgs) to investigate how human hepatocytes initiate and execute endomitosis. We find that cells in endomitosis M phase have normal mitotic timings, but lose membrane anchorage to the midbody during cytokinesis, which is associated with the loss of four cortical anchoring proteins, RacGAP1, Anillin, SEPT9, and citron kinase (CIT-K). Moreover, reduction of WNT activity increases the percentage of binucleated cells in Hep-Orgs, an effect that is dependent on the atypical E2F proteins, E2F7 and E2F8. Together, we have elucidated how hepatocytes undergo endomitosis in human Hep-Orgs, providing new insights into the mechanisms of endomitosis in mammals.
© 2024 Darmasaputra et al.
Conflict of interest statement
Disclosures: H. Clevers reported personal fees from F. Hoffmann-La Roche Ltd., Basel, Switzerland, outside the submitted work; in addition, H. Clevers had a patent to Application PCT/EP2019/082618 events 2019-11-26 Application filed by Koninklijke Nederlandse Akademie Van Wetenschappen 2019-11-26 Priority to US17/296,049 2020-06-04 Publication of WO2020109324A1 licensed “HUB, Utrecht, NL”; and “I am currently an employee of F. Hoffmann-La Roche Ltd. in Basel, Switzerland, where I am member of the Extended Executive Board and head Pharma Research & Early Development. The company has no involvement in the work published in this manuscript.” No other disclosures were reported.
Figures
References
-
- Artegiani, B., Hendriks D., Beumer J., Kok R., Zheng X., Joore I., Chuva de Sousa Lopes S., van Zon J., Tans S., and Clevers H.. 2020. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol. 22:321–331. 10.1038/s41556-020-0472-5 - DOI - PubMed
-
- Bodor, D. 2021. Mitotic scoring tool: version 0.1.0. github.com/DaniBodor/MitoticScoring.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

