Mechanics limits ecological diversity and promotes heterogeneity in confined bacterial communities
- PMID: 38728226
- PMCID: PMC11098131
- DOI: 10.1073/pnas.2322321121
Mechanics limits ecological diversity and promotes heterogeneity in confined bacterial communities
Abstract
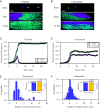
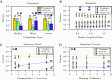
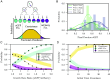
Multispecies bacterial populations often inhabit confined and densely packed environments where spatial competition determines the ecological diversity of the community. However, the role of mechanical interactions in shaping the ecology is still poorly understood. Here, we study a model system consisting of two populations of nonmotile Escherichia coli bacteria competing within open, monolayer microchannels. The competitive dynamics is observed to be biphasic: After seeding, either one strain rapidly fixates or both strains orient into spatially stratified, stable communities. We find that mechanical interactions with other cells and local spatial constraints influence the resulting community ecology in unexpected ways, severely limiting the overall diversity of the communities while simultaneously allowing for the establishment of stable, heterogeneous populations of bacteria displaying disparate growth rates. Surprisingly, the populations have a high probability of coexisting even when one strain has a significant growth advantage. A more coccus morphology is shown to provide a selective advantage, but agent-based simulations indicate this is due to hydrodynamic and adhesion effects within the microchannel and not from breaking of the nematic ordering. Our observations are qualitatively reproduced by a simple Pólya urn model, which suggests the generality of our findings for confined population dynamics and highlights the importance of early colonization conditions on the resulting diversity and ecology of bacterial communities. These results provide fundamental insights into the determinants of community diversity in dense confined ecosystems where spatial exclusion is central to competition as in organized biofilms or intestinal crypts.
Keywords: microbial ecology; microbial systems; physical biology; population dynamics; quantitative biology.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Stoodley P., Sauer K., Davies D. G., Costerton J. W., Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002). - PubMed
-
- Flemming H. C., et al. , Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016). - PubMed
-
- Shapiro J. A., Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104 (1998). - PubMed
-
- Nadell C. D., Drescher K., Foster K. R., Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
- RGPIN-2019-06520/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- RGPIN-2022-04909/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- 207-2019-2020-Q4-00576/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
LinkOut - more resources
Full Text Sources

