The future of rapid and automated single-cell data analysis using reference mapping
- PMID: 38729109
- PMCID: PMC11184658
- DOI: 10.1016/j.cell.2024.03.009
The future of rapid and automated single-cell data analysis using reference mapping
Abstract
As the number of single-cell datasets continues to grow rapidly, workflows that map new data to well-curated reference atlases offer enormous promise for the biological community. In this perspective, we discuss key computational challenges and opportunities for single-cell reference-mapping algorithms. We discuss how mapping algorithms will enable the integration of diverse datasets across disease states, molecular modalities, genetic perturbations, and diverse species and will eventually replace manual and laborious unsupervised clustering pipelines.
Keywords: cross-species comparisons; machine learning; multimodal analysis; reference mapping; single-cell analysis.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.L. consults Santa Ana Bio, worked as a part-time employee at Relation Therapeutics, and owns interests in Relation Therapeutics and is a part-time employee at AIVIVO. F.J.T. consults for Immunai Inc., CytoReason Ltd, Cellarity Inc, and Omniscope Ltd, and owns interests in Dermagnostix GmbH and Cellarity Inc. In the past 3 years, R.S. has worked as a consultant for Bristol-Myers Squibb, Regeneron, and Kallyope and served as a scientific advisory board member for ImmunAI, Resolve Biosciences, Nanostring, and the NYC Pandemic Response Lab. R.S. and Y.H. are co-founders and equity holders of Neptune Bio. As of August 1, 2023, Y.H. is an employee of Neptune Bio.
Figures


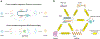
References
-
- Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

