Understanding the complexity of p53 in a new era of tumor suppression
- PMID: 38729160
- PMCID: PMC11190820
- DOI: 10.1016/j.ccell.2024.04.009
Understanding the complexity of p53 in a new era of tumor suppression
Abstract
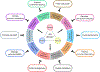
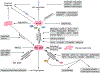
p53 was discovered 45 years ago as an SV40 large T antigen binding protein, coded by the most frequently mutated TP53 gene in human cancers. As a transcription factor, p53 is tightly regulated by a rich network of post-translational modifications to execute its diverse functions in tumor suppression. Although early studies established p53-mediated cell-cycle arrest, apoptosis, and senescence as the classic barriers in cancer development, a growing number of new functions of p53 have been discovered and the scope of p53-mediated anti-tumor activity is largely expanded. Here, we review the complexity of different layers of p53 regulation, and the recent advance of the p53 pathway in metabolism, ferroptosis, immunity, and others that contribute to tumor suppression. We also discuss the challenge regarding how to activate p53 function specifically effective in inhibiting tumor growth without harming normal homeostasis for cancer therapy.
Keywords: MDM2; MDMX; apoptosis; cancer treatment; cell competition; cell-cycle arrest; ferroptosis; genome stability; immunity; metabolism; metastasis; p53; p53 mutation; p63; p73; senescence; stem cell dynamics; targeting p53; tumor suppression.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests O.T. is currently an employee of AstraZeneca and has stock ownership in AstraZeneca.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

