Assessment of three antibiotic combination regimens against Gram-negative bacteria causing neonatal sepsis in low- and middle-income countries
- PMID: 38729951
- PMCID: PMC11087563
- DOI: 10.1038/s41467-024-48296-z
Assessment of three antibiotic combination regimens against Gram-negative bacteria causing neonatal sepsis in low- and middle-income countries
Abstract
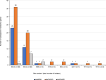
Gram-negative bacteria (GNB) are a major cause of neonatal sepsis in low- and middle-income countries (LMICs). Although the World Health Organization (WHO) reports that over 80% of these sepsis deaths could be prevented through improved treatment, the efficacy of the currently recommended first- and second-line treatment regimens for this condition is increasingly affected by high rates of drug resistance. Here we assess three well known antibiotics, fosfomycin, flomoxef and amikacin, in combination as potential antibiotic treatment regimens by investigating the drug resistance and genetic profiles of commonly isolated GNB causing neonatal sepsis in LMICs. The five most prevalent bacterial isolates in the NeoOBS study (NCT03721302) are Klebsiella pneumoniae, Acinetobacter baumannii, E. coli, Serratia marcescens and Enterobacter cloacae complex. Among these isolates, high levels of ESBL and carbapenemase encoding genes are detected along with resistance to ampicillin, gentamicin and cefotaxime, the current WHO recommended empiric regimens. The three new combinations show excellent in vitro activity against ESBL-producing K. pneumoniae and E. coli isolates. Our data should further inform and support the clinical evaluation of these three antibiotic combinations for the treatment of neonatal sepsis in areas with high rates of multidrug-resistant Gram-negative bacteria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: J.A.B.: Research grant support to the university from Wellcome, NIHR, MRC, and the Bill & Melinda Gates Foundation who had no role in any aspect of the study or decision to publish. A.S.W. is an NIHR Senior Investigator supported by the NIHR Biomedical Research Center Oxford and core support to the MRC Clinical Trials Unit [MC_UU_00004/05]. J.B. is an NIHR Advanced Fellow and Chief Investigator supported by grant NIHR302554 and H2020 Agreement number 965328. The remaining authors declare no competing interests.
Figures





References
-
- World Health Organization. Global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions. https://www.who.int/publications/i/item/9789240010789. (2020) (Last access date November 1, 2023).
-
- Investigators of the Delhi Neonatal Infection Study (DeNIS) collaboration Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Glob. Health. 2016;4:e752–e760. doi: 10.1016/S2214-109X(16)30148-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

