Assessing the role of programmed cell death signatures and related gene TOP2A in progression and prognostic prediction of clear cell renal cell carcinoma
- PMID: 38730293
- PMCID: PMC11084013
- DOI: 10.1186/s12935-024-03346-w
Assessing the role of programmed cell death signatures and related gene TOP2A in progression and prognostic prediction of clear cell renal cell carcinoma
Abstract
Kidney Clear Cell Carcinoma (KIRC), the predominant form of kidney cancer, exhibits a diverse therapeutic response to Immune Checkpoint Inhibitors (ICIs), highlighting the need for predictive models of ICI efficacy. Our study has constructed a prognostic model based on 13 types of Programmed Cell Death (PCD), which are intertwined with tumor progression and the immune microenvironment. Validated by analyses of comprehensive datasets, this model identifies seven key PCD genes that delineate two subtypes with distinct immune profiles and sensitivities to anti-PD-1 therapy. The high-PCD group demonstrates a more immune-suppressive environment, while the low-PCD group shows better responses to PD-1 treatment. In particular, TOP2A emerged as crucial, with its inhibition markedly reducing KIRC cell growth and mobility. These findings underscore the relevance of PCDs in predicting KIRC outcomes and immunotherapy response, with implications for enhancing clinical decision-making.
Keywords: Immune microenvironment; Immunotherapy; KIRC; PCD; TOP2A.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
The authors declare no conflict of interest for this article.
Figures
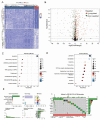



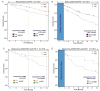

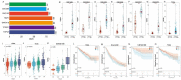

References
-
- Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MT, Venugopal B, Kollmannsberger C, Negrier S, Uemura M, Lee JL, Vasiliev A, Miller WH, Jr, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2019;380:1103–15. doi: 10.1056/NEJMoa1816047. - DOI - PMC - PubMed
-
- Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov D, Pouliot F, Alekseev B, Soulières D, Melichar B, Vynnychenko I, Kryzhanivska A, Bondarenko I, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2019;380:1116–27. doi: 10.1056/NEJMoa1816714. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

