Multiomics-based molecular subtyping based on the commensal microbiome predicts molecular characteristics and the therapeutic response in breast cancer
- PMID: 38730464
- PMCID: PMC11083817
- DOI: 10.1186/s12943-024-02017-8
Multiomics-based molecular subtyping based on the commensal microbiome predicts molecular characteristics and the therapeutic response in breast cancer
Abstract
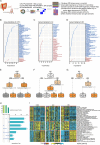
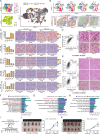
The gut microbiota has been demonstrated to be correlated with the clinical phenotypes of diseases, including cancers. However, there are few studies on clinical subtyping based on the gut microbiota, especially in breast cancer (BC) patients. Here, using machine learning methods, we analysed the gut microbiota of BC, colorectal cancer (CRC), and gastric cancer (GC) patients to identify their shared metabolic pathways and the importance of these pathways in cancer development. Based on the gut microbiota-related metabolic pathways, human gene expression profile and patient prognosis, we established a novel BC subtyping system and identified a subtype called "challenging BC". Tumours with this subtype have more genetic mutations and a more complex immune environment than those of other subtypes. A score index was proposed for in-depth analysis and showed a significant negative correlation with patient prognosis. Notably, activation of the TPK1-FOXP3-mediated Hedgehog signalling pathway and TPK1-ITGAE-mediated mTOR signalling pathway was linked to poor prognosis in "challenging BC" patients with high scores, as validated in a patient-derived xenograft (PDX) model. Furthermore, our subtyping system and score index are effective predictors of the response to current neoadjuvant therapy regimens, with the score index significantly negatively correlated with both treatment efficacy and the number of immune cells. Therefore, our findings provide valuable insights into predicting molecular characteristics and treatment responses in "challenging BC" patients.
© 2024. The Author(s).
Conflict of interest statement
All authors declare no competing interests associated with this study.
Figures




References
-
- Yu J, et al. Bifidobacterium longum promotes postoperative liver function recovery in patients with hepatocellular carcinoma. Cell Host Microbe; 2023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

