Pioglitazone Phases and Metabolic Effects in Nanoparticle-Treated Cells Analyzed via Rapid Visualization of FLIM Images
- PMID: 38731628
- PMCID: PMC11085555
- DOI: 10.3390/molecules29092137
Pioglitazone Phases and Metabolic Effects in Nanoparticle-Treated Cells Analyzed via Rapid Visualization of FLIM Images
Abstract
Fluorescence lifetime imaging microscopy (FLIM) has proven to be a useful method for analyzing various aspects of material science and biology, like the supramolecular organization of (slightly) fluorescent compounds or the metabolic activity in non-labeled cells; in particular, FLIM phasor analysis (phasor-FLIM) has the potential for an intuitive representation of complex fluorescence decays and therefore of the analyzed properties. Here we present and make available tools to fully exploit this potential, in particular by coding via hue, saturation, and intensity the phasor positions and their weights both in the phasor plot and in the microscope image. We apply these tools to analyze FLIM data acquired via two-photon microscopy to visualize: (i) different phases of the drug pioglitazone (PGZ) in solutions and/or crystals, (ii) the position in the phasor plot of non-labelled poly(lactic-co-glycolic acid) (PLGA) nanoparticles (NPs), and (iii) the effect of PGZ or PGZ-containing NPs on the metabolism of insulinoma (INS-1 E) model cells. PGZ is recognized for its efficacy in addressing insulin resistance and hyperglycemia in type 2 diabetes mellitus, and polymeric nanoparticles offer versatile platforms for drug delivery due to their biocompatibility and controlled release kinetics. This study lays the foundation for a better understanding via phasor-FLIM of the organization and effects of drugs, in particular, PGZ, within NPs, aiming at better control of encapsulation and pharmacokinetics, and potentially at novel anti-diabetics theragnostic nanotools.
Keywords: MATLAB tool; PLGA nanoparticles; drug delivery; fluorescence lifetime imaging microscopy (FLIM); insulinoma (INS-1) cells; phasor-FLIM analysis; pioglitazone characterization.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


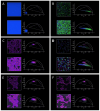
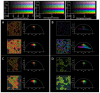
References
-
- Vitali M., Picazo F., Prokazov Y., Duci A., Turbin E., Götze C., Llopis J., Hartig R., Visser A.J.W.G., Zuschratter W. Wide-Field Multi-Parameter FLIM: Long-Term Minimal Invasive Observation of Proteins in Living Cells. PLoS ONE. 2011;6:e15820. doi: 10.1371/journal.pone.0015820. - DOI - PMC - PubMed
-
- Stringari C., Cinquin A., Cinquin O., Digman M.A., Donovan P.J., Gratton E. Phasor Approach to Fluorescence Lifetime Microscopy Distinguishes Different Metabolic States of Germ Cells in a Live Tissue. Proc. Natl. Acad. Sci. USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. - DOI - PMC - PubMed
-
- Bec J., Vela D., Phipps J.E., Agung M., Unger J., Margulies K.B., Southard J.A., Buja L.M., Marcu L. Label-Free Visualization and Quantification of Biochemical Markers of Atherosclerotic Plaque Progression Using Intravascular Fluorescence Lifetime. JACC Cardiovasc. Imaging. 2021;14:1832–1842. doi: 10.1016/j.jcmg.2020.10.004. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

