Establishment of a Simple, Sensitive, and Specific Salmonella Detection Method Based on Recombinase-Aided Amplification Combined with dsDNA-Specific Nucleases
- PMID: 38731750
- PMCID: PMC11083397
- DOI: 10.3390/foods13091380
Establishment of a Simple, Sensitive, and Specific Salmonella Detection Method Based on Recombinase-Aided Amplification Combined with dsDNA-Specific Nucleases
Abstract
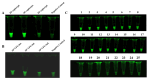
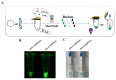
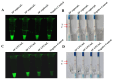
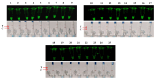
Salmonella is a common foodborne pathogen that can cause food poisoning, posing a serious threat to human health. Therefore, quickly, sensitively, and accurately detecting Salmonella is crucial to ensuring food safety. For the Salmonella hilA gene, we designed Recombinase-aided amplification (RAA) primers and dsDNA-specific nuclease (DNase) probes. The ideal primer and probe combination was found when conditions were optimized. Under UV light, a visual Salmonella detection technique (RAA-dsDNase) was developed. Additionally, the RAA-dsDNase was modified to further reduce pollution hazards and simplify operations. One-pot RAA-dsDNase-UV or one-pot RAA-dsDNase-LFD was developed as a Salmonella detection method, using UV or a lateral flow dipstick (LFD) for result observation. Among them, one-pot RAA-dsDNase and one-pot RAA-dsDNase-LFD had detection times of 50 min and 60 min, respectively, for detecting Salmonella genomic DNA. One-pot RAA-dsDNase-UV had a detection limit of 101 copies/μL and 101 CFU/mL, while one-pot RAA-dsDNase-LFD had a sensitivity of 102 copies/μL and 102 CFU/mL. One-pot RAA-dsDNase-UV and one-pot RAA-dsDNase-LFD assays may identify 17 specific Salmonella serovars witho ut causing a cross-reaction with the remaining 8 bacteria, which include E. coli. Furthermore, Salmonella in tissue and milk samples has been reliably detected using both approaches. Overall, the detection method developed in this study can quickly, sensitively, and accurately detect Salmonella, and it is expected to become an important detection tool for the prevention and control of Salmonella in the future.
Keywords: RAA; Salmonella; dsDNase; one pot; visualization.
Conflict of interest statement
The author declares no competing interests.
Figures

Similar articles
-
Establishment of reverse transcription recombinase-aided amplification-lateral-flow dipstick and real-time fluorescence-based reverse transcription recombinase-aided amplification methods for detection of the Newcastle disease virus in chickens.Poult Sci. 2020 Jul;99(7):3393-3401. doi: 10.1016/j.psj.2020.03.018. Epub 2020 Apr 15. Poult Sci. 2020. PMID: 32616233 Free PMC article.
-
Establishment and Application of Duplex Recombinase-Aided Amplification Combined with Lateral Flow Dipsticks for Rapid and Simultaneous Visual Detection of Klebsiella pneumoniae and Staphylococcus aureus in Milk.Foods. 2025 Feb 9;14(4):573. doi: 10.3390/foods14040573. Foods. 2025. PMID: 40002017 Free PMC article.
-
Rapid and visual detection of Mycoplasma synoviae by recombinase-aided amplification assay combined with a lateral flow dipstick.Poult Sci. 2022 Jul;101(7):101860. doi: 10.1016/j.psj.2022.101860. Epub 2022 Mar 15. Poult Sci. 2022. PMID: 35537343 Free PMC article.
-
Development of a recombinase-aided amplification method combined with lateral flow dipstick assay to detect Porcine circovirus type 2.Vet World. 2023 Nov;16(11):2313-2320. doi: 10.14202/vetworld.2023.2313-2320. Epub 2023 Nov 19. Vet World. 2023. PMID: 38152256 Free PMC article.
-
Development of two recombinase-aided amplification assays combined with lateral flow dipstick (RAA-LFD) and real-time fluorescence (RF-RAA) for the detection of Frog virus 3-like ranaviruses.Fish Shellfish Immunol. 2025 Mar;158:110154. doi: 10.1016/j.fsi.2025.110154. Epub 2025 Jan 23. Fish Shellfish Immunol. 2025. PMID: 39863138
Cited by
-
Molecular methods for rapid detection and identification of foodborne pathogenic bacteria.World J Microbiol Biotechnol. 2025 May 15;41(5):175. doi: 10.1007/s11274-025-04396-6. World J Microbiol Biotechnol. 2025. PMID: 40369382 Review.
-
A rapid and visual detection assay for Senecavirus A based on recombinase-aided amplification and lateral flow dipstick.Front Cell Infect Microbiol. 2024 Oct 23;14:1474676. doi: 10.3389/fcimb.2024.1474676. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39507945 Free PMC article.
References
-
- Xue L., Guo R., Huang F., Qi W., Liu Y., Cai G., Lin J. An impedance biosensor based on magnetic nanobead net and MnO2 nanoflowers for rapid and sensitive detection of foodborne bacteria. Biosens. Bioelectron. 2020;173:112800. - PubMed
-
- Delibato E., Volpe G., Romanazzo D., De Medici D., Toti L., Moscone D., Palleschi G. Development and application of an electrochemical plate coupled with immunomagnetic beads (ELIME) array for Salmonella enterica detection in meat samples. J. Agric. Food Chem. 2009;57:7200–7204. doi: 10.1021/jf901181m. - DOI - PubMed
-
- Jarvis N.A., O’Bryan C.A., Dawoud T.M., Park S.H., Kwon Y.M., Crandall P.G., Ricke S.C. An overview of Salmonella thermal destruction during food processing and preparation. Food Control. 2016;68:280–290. doi: 10.1016/j.foodcont.2016.04.006. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

