Variable Penetrance and Expressivity of a Rare Pore Loss-of-Function Mutation (p.L889V) of Nav1.5 Channels in Three Spanish Families
- PMID: 38731905
- PMCID: PMC11083067
- DOI: 10.3390/ijms25094686
Variable Penetrance and Expressivity of a Rare Pore Loss-of-Function Mutation (p.L889V) of Nav1.5 Channels in Three Spanish Families
Abstract
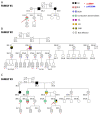
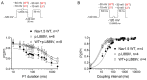
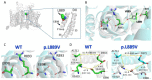
A novel rare mutation in the pore region of Nav1.5 channels (p.L889V) has been found in three unrelated Spanish families that produces quite diverse phenotypic manifestations (Brugada syndrome, conduction disease, dilated cardiomyopathy, sinus node dysfunction, etc.) with variable penetrance among families. We clinically characterized the carriers and recorded the Na+ current (INa) generated by p.L889V and native (WT) Nav1.5 channels, alone or in combination, to obtain further insight into the genotypic-phenotypic relationships in patients carrying SCN5A mutations and in the molecular determinants of the Nav1.5 channel function. The variant produced a strong dominant negative effect (DNE) since the peak INa generated by p.L889V channels expressed in Chinese hamster ovary cells, either alone (-69.4 ± 9.0 pA/pF) or in combination with WT (-62.2 ± 14.6 pA/pF), was significantly (n ≥ 17, p < 0.05) reduced compared to that generated by WT channels alone (-199.1 ± 44.1 pA/pF). The mutation shifted the voltage dependence of channel activation and inactivation to depolarized potentials, did not modify the density of the late component of INa, slightly decreased the peak window current, accelerated the recovery from fast and slow inactivation, and slowed the induction kinetics of slow inactivation, decreasing the fraction of channels entering this inactivated state. The membrane expression of p.L889V channels was low, and in silico molecular experiments demonstrated profound alterations in the disposition of the pore region of the mutated channels. Despite the mutation producing a marked DNE and reduction in the INa and being located in a critical domain of the channel, its penetrance and expressivity are quite variable among the carriers. Our results reinforce the argument that the incomplete penetrance and phenotypic variability of SCN5A loss-of-function mutations are the result of a combination of multiple factors, making it difficult to predict their expressivity in the carriers despite the combination of clinical, genetic, and functional studies.
Keywords: Brugada syndrome; Nav1.5; SCN5A; cardiac conduction defect; dilated cardiomyopathy; mutation; phenotypic penetrance; phenotypic variability.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Remme C.A., Verkerk A.O., Hoogaars W.M.H., Aanhaanen W.T.J., Scicluna B.P., Annink C., van den Hoff M.J.B., Wilde A.A., van Veen T.A.B., Veldkamp M.W., et al. The Cardiac Sodium Channel Displays Differential Distribution in the Conduction System and Transmural Heterogeneity in the Murine Ventricular Myocardium. Basic Res. Cardiol. 2009;104:511–522. doi: 10.1007/s00395-009-0012-8. - DOI - PMC - PubMed
-
- Remme C.A., Verkerk A.O., Nuyens D., van Ginneken A.C.G., van Brunschot S., Belterman C.N.W., Wilders R., van Roon M.A., Tan H.L., Wilde A.A.M., et al. Overlap Syndrome of Cardiac Sodium Channel Disease in Mice Carrying the Equivalent Mutation of Human SCN5A-1795insD. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

