Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942
- PMID: 38731921
- PMCID: PMC11083307
- DOI: 10.3390/ijms25094702
Analysing the Cyanobacterial PipX Interaction Network Using NanoBiT Complementation in Synechococcus elongatus PCC7942
Abstract
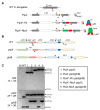
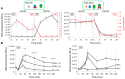
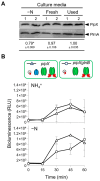
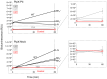
The conserved cyanobacterial protein PipX is part of a complex interaction network with regulators involved in essential processes that include metabolic homeostasis and ribosome assembly. Because PipX interactions depend on the relative levels of their different partners and of the effector molecules binding to them, in vivo studies are required to understand the physiological significance and contribution of environmental factors to the regulation of PipX complexes. Here, we have used the NanoBiT complementation system to analyse the regulation of complex formation in Synechococcus elongatus PCC 7942 between PipX and each of its two best-characterized partners, PII and NtcA. Our results confirm previous in vitro analyses on the regulation of PipX-PII and PipX-NtcA complexes by 2-oxoglutarate and on the regulation of PipX-PII by the ATP/ADP ratio, showing the disruption of PipX-NtcA complexes due to increased levels of ADP-bound PII in Synechococcus elongatus. The demonstration of a positive role of PII on PipX-NtcA complexes during their initial response to nitrogen starvation or the impact of a PipX point mutation on the activity of PipX-PII and PipX-NtcA reporters are further indications of the sensitivity of the system. This study reveals additional regulatory complexities in the PipX interaction network, opening a path for future research on cyanobacteria.
Keywords: 2-oxoglutarate; NanoLuc; NtcA; PCAs; PII; complementation reporter; energy regulation; environmental factors; nitrogen regulation; protein-fragment complementation assays.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA.Mol Microbiol. 2006 Jul;61(2):457-69. doi: 10.1111/j.1365-2958.2006.05231.x. Epub 2006 Jun 1. Mol Microbiol. 2006. PMID: 16796668
-
PipX, the coactivator of NtcA, is a global regulator in cyanobacteria.Proc Natl Acad Sci U S A. 2014 Jun 10;111(23):E2423-30. doi: 10.1073/pnas.1404097111. Epub 2014 May 27. Proc Natl Acad Sci U S A. 2014. PMID: 24912181 Free PMC article.
-
Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII.Proc Natl Acad Sci U S A. 2010 Aug 31;107(35):15397-402. doi: 10.1073/pnas.1007015107. Epub 2010 Aug 17. Proc Natl Acad Sci U S A. 2010. PMID: 20716687 Free PMC article.
-
The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-covalent Interactions.Front Mol Biosci. 2018 Nov 13;5:91. doi: 10.3389/fmolb.2018.00091. eCollection 2018. Front Mol Biosci. 2018. PMID: 30483512 Free PMC article. Review.
-
Distinctive Features of PipX, a Unique Signaling Protein of Cyanobacteria.Life (Basel). 2020 May 28;10(6):79. doi: 10.3390/life10060079. Life (Basel). 2020. PMID: 32481703 Free PMC article. Review.
Cited by
-
Structures of the cyanobacterial nitrogen regulators NtcA and PipX complexed to DNA shed light on DNA binding by NtcA and implicate PipX in the recruitment of RNA polymerase.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf096. doi: 10.1093/nar/gkaf096. Nucleic Acids Res. 2025. PMID: 39995039 Free PMC article.
-
Studies on the PII-PipX-NtcA Regulatory Axis of Cyanobacteria Provide Novel Insights into the Advantages and Limitations of Two-Hybrid Systems for Protein Interactions.Int J Mol Sci. 2024 May 16;25(10):5429. doi: 10.3390/ijms25105429. Int J Mol Sci. 2024. PMID: 38791467 Free PMC article.
References
-
- Lee H.-W., Noh J.-H., Choi D.-H., Yun M., Bhavya P.S., Kang J.-J., Lee J.-H., Kim K.-W., Jang H.-K., Lee S.-H. Picocyanobacterial Contribution to the Total Primary Production in the Northwestern Pacific Ocean. Water. 2021;13:1610. doi: 10.3390/w13111610. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

