Development, High-Throughput Profiling, and Biopanning of a Large Phage Display Single-Domain Antibody Library
- PMID: 38732011
- PMCID: PMC11083953
- DOI: 10.3390/ijms25094791
Development, High-Throughput Profiling, and Biopanning of a Large Phage Display Single-Domain Antibody Library
Abstract
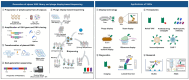
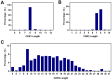
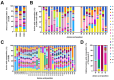
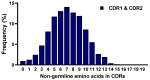
Immunoglobulin G-based monoclonal antibodies (mAbs) have been effective in treating various diseases, but their large molecular size can limit their penetration of tissue and efficacy in multifactorial diseases, necessitating the exploration of alternative forms. In this study, we constructed a phage display library comprising single-domain antibodies (sdAbs; or "VHHs"), known for their small size and remarkable stability, using a total of 1.6 × 109 lymphocytes collected from 20 different alpacas, resulting in approximately 7.16 × 1010 colonies. To assess the quality of the constructed library, next-generation sequencing-based high-throughput profiling was performed, analyzing approximately 5.65 × 106 full-length VHH sequences, revealing 92% uniqueness and confirming the library's diverse composition. Systematic characterization of the library revealed multiple sdAbs with high affinity for three therapeutically relevant antigens. In conclusion, our alpaca sdAb phage display library provides a versatile resource for diagnostics and therapeutics. Furthermore, the library's vast natural VHH antibody repertoire offers insights for generating humanized synthetic sdAb libraries, further advancing sdAb-based therapeutics.
Keywords: antibody library; next-generation sequencing; phage display; single-domain antibody.
Conflict of interest statement
Dr. Taehoon Ryu and Yushin Jung were employed by ATG Lifetech Inc. The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Joining the in vitro immunization of alpaca lymphocytes and phage display: rapid and cost effective pipeline for sdAb synthesis.Microb Cell Fact. 2017 Jan 23;16(1):13. doi: 10.1186/s12934-017-0630-z. Microb Cell Fact. 2017. PMID: 28114943 Free PMC article.
-
Oxford nanopore sequencing enables rapid discovery of single-domain antibodies from phage display libraries.Biotechniques. 2018 Dec;65(6):351-356. doi: 10.2144/btn-2018-0123. Biotechniques. 2018. PMID: 30477332
-
Facile Affinity Maturation of Single-Domain Antibodies Using Next-Generation DNA Sequencing.Methods Mol Biol. 2022;2446:245-268. doi: 10.1007/978-1-0716-2075-5_12. Methods Mol Biol. 2022. PMID: 35157277
-
Cognizance of Molecular Methods for the Generation of Mutagenic Phage Display Antibody Libraries for Affinity Maturation.Int J Mol Sci. 2019 Apr 15;20(8):1861. doi: 10.3390/ijms20081861. Int J Mol Sci. 2019. PMID: 30991723 Free PMC article. Review.
-
Infectious disease antibodies for biomedical applications: A mini review of immune antibody phage library repertoire.Int J Biol Macromol. 2020 Nov 15;163:640-648. doi: 10.1016/j.ijbiomac.2020.06.268. Epub 2020 Jul 8. Int J Biol Macromol. 2020. PMID: 32650013 Free PMC article. Review.
Cited by
-
High-throughput strategies for monoclonal antibody screening: advances and challenges.J Biol Eng. 2025 May 8;19(1):41. doi: 10.1186/s13036-025-00513-z. J Biol Eng. 2025. PMID: 40340930 Free PMC article. Review.
-
Unveiling the new chapter in nanobody engineering: advances in traditional construction and AI-driven optimization.J Nanobiotechnology. 2025 Feb 6;23(1):87. doi: 10.1186/s12951-025-03169-5. J Nanobiotechnology. 2025. PMID: 39915791 Free PMC article. Review.
References
-
- Quinteros D.A., Bermúdez J.M., Ravetti S., Cid A., Allemandi D.A., Palma S.D. Nanostructures for Drug Delivery. Elsevier; Amsterdam, The Netherlands: 2017. Chapter 25—Therapeutic use of monoclonal antibodies: General aspects and challenges for drug delivery; pp. 807–833.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

