Structural basis of human NOX5 activation
- PMID: 38734761
- PMCID: PMC11088703
- DOI: 10.1038/s41467-024-48467-y
Structural basis of human NOX5 activation
Abstract
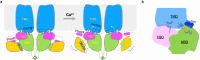
NADPH oxidase 5 (NOX5) catalyzes the production of superoxide free radicals and regulates physiological processes from sperm motility to cardiac rhythm. Overexpression of NOX5 leads to cancers, diabetes, and cardiovascular diseases. NOX5 is activated by intracellular calcium signaling, but the underlying molecular mechanism of which - in particular, how calcium triggers electron transfer from NADPH to FAD - is still unclear. Here we capture motions of full-length human NOX5 upon calcium binding using single-particle cryogenic electron microscopy (cryo-EM). By combining biochemistry, mutagenesis analyses, and molecular dynamics (MD) simulations, we decode the molecular basis of NOX5 activation and electron transfer. We find that calcium binding to the EF-hand domain increases NADPH dynamics, permitting electron transfer between NADPH and FAD and superoxide production. Our structural findings also uncover a zinc-binding motif that is important for NOX5 stability and enzymatic activity, revealing modulation mechanisms of reactive oxygen species (ROS) production.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

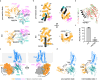


References
-
- Devasagayam TP, et al. Free radicals and antioxidants in human health: current status and future prospects. J Assoc. Physicians India. 2004;52:794–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

