Neuropathy target esterase activity defines phenotypes among PNPLA6 disorders
- PMID: 38735647
- PMCID: PMC11146429
- DOI: 10.1093/brain/awae055
Neuropathy target esterase activity defines phenotypes among PNPLA6 disorders
Abstract
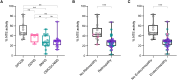
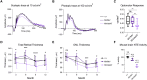
Biallelic pathogenic variants in the PNPLA6 gene cause a broad spectrum of disorders leading to gait disturbance, visual impairment, anterior hypopituitarism and hair anomalies. PNPLA6 encodes neuropathy target esterase (NTE), yet the role of NTE dysfunction on affected tissues in the large spectrum of associated disease remains unclear. We present a systematic evidence-based review of a novel cohort of 23 new patients along with 95 reported individuals with PNPLA6 variants that implicate missense variants as a driver of disease pathogenesis. Measuring esterase activity of 46 disease-associated and 20 common variants observed across PNPLA6-associated clinical diagnoses unambiguously reclassified 36 variants as pathogenic and 10 variants as likely pathogenic, establishing a robust functional assay for classifying PNPLA6 variants of unknown significance. Estimating the overall NTE activity of affected individuals revealed a striking inverse relationship between NTE activity and the presence of retinopathy and endocrinopathy. This phenomenon was recaptured in vivo in an allelic mouse series, where a similar NTE threshold for retinopathy exists. Thus, PNPLA6 disorders, previously considered allelic, are a continuous spectrum of pleiotropic phenotypes defined by an NTE genotype:activity:phenotype relationship. This relationship, and the generation of a preclinical animal model, pave the way for therapeutic trials, using NTE as a biomarker.
Keywords: Boucher-Neuhäuser syndrome; Gordon-Holmes syndrome; Laurence-Moon syndrome; Oliver-McFarlane syndrome; neurodegeneration; spastic paraplegia type 39.
Published by Oxford University Press on behalf of the Guarantors of Brain 2024.
Conflict of interest statement
The authors report no competing interests.
Figures



Update of
-
Neuropathy target esterase activity predicts retinopathy among PNPLA6 disorders.bioRxiv [Preprint]. 2023 Jun 11:2023.06.09.544373. doi: 10.1101/2023.06.09.544373. bioRxiv. 2023. Update in: Brain. 2024 Jun 3;147(6):2085-2097. doi: 10.1093/brain/awae055. PMID: 37333224 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

