Throat microbiota drives alterations in pulmonary alveolar microbiota in patients with septic ARDS
- PMID: 38736041
- PMCID: PMC11093027
- DOI: 10.1080/21505594.2024.2350775
Throat microbiota drives alterations in pulmonary alveolar microbiota in patients with septic ARDS
Abstract
Objectives: The translocation of intestinal flora has been linked to the colonization of diverse and heavy lower respiratory flora in patients with septic ARDS, and is considered a critical prognostic factor for patients.
Methods: On the first and third days of ICU admission, BALF, throat swab, and anal swab were collected, resulting in a total of 288 samples. These samples were analyzed using 16S rRNA analysis and the traceability analysis of new generation technology.
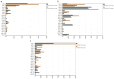
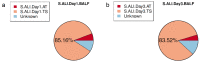
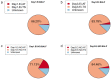
Results: On the first day, among the top five microbiota species in abundance, four species were found to be identical in BALF and throat samples. Similarly, on the third day, three microbiota species were found to be identical in abundance in both BALF and throat samples. On the first day, 85.16% of microorganisms originated from the throat, 5.79% from the intestines, and 9.05% were unknown. On the third day, 83.52% of microorganisms came from the throat, 4.67% from the intestines, and 11.81% were unknown. Additionally, when regrouping the 46 patients, the results revealed a significant predominance of throat microorganisms in BALF on both the first and third day. Furthermore, as the disease progressed, the proportion of intestinal flora in BALF increased in patients with enterogenic ARDS.
Conclusions: In patients with septic ARDS, the main source of lung microbiota is primarily from the throat. Furthermore, the dynamic trend of the microbiota on the first and third day is essentially consistent.It is important to note that the origin of the intestinal flora does not exclude the possibility of its origin from the throat.
Keywords: 16S rRNA; ARDS; Sepsis; anal swab; bronchoalveolar lavage fluid; throat swab.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
