Molecular and immune characterization of Chinese early-stage non-squamous non-small cell lung cancer: a multi-omics cohort study
- PMID: 38736486
- PMCID: PMC11082711
- DOI: 10.21037/tlcr-23-800
Molecular and immune characterization of Chinese early-stage non-squamous non-small cell lung cancer: a multi-omics cohort study
Abstract
Background: Albeit considered with superior survival, around 30% of the early-stage non-squamous non-small cell lung cancer (Ns-NSCLC) patients relapse within 5 years, suggesting unique biology. However, the biological characteristics of early-stage Ns-NSCLC, especially in the Chinese population, are still unclear.
Methods: Multi-omics interrogation of early-stage Ns-NSCLC (stage I-III), paired blood samples and normal lung tissues (n=76) by whole-exome sequencing (WES), RNA sequencing, and T-cell receptor (TCR) sequencing were conducted.
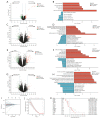
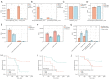
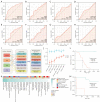
Results: An average of 128 exonic mutations were identified, and the most frequently mutant gene was EGFR (55%), followed by TP53 (37%) and TTN (26%). Mutations in MUC17, ABCA2, PDE4DIP, and MYO18B predicted significantly unfavorable disease-free survival (DFS). Moreover, cytobands amplifications in 8q24.3, 14q13.1, 14q11.2, and deletion in 3p21.1 were highlighted in recurrent cases. Higher incidence of human leukocyte antigen loss of heterozygosity (HLA-LOH), higher tumor mutational burden (TMB) and tumor neoantigen burden (TNB) were identified in ever-smokers than never-smokers. HLA-LOH also correlated with higher TMB, TNB, intratumoral heterogeneity (ITH), and whole chromosomal instability (wCIN) scores. Interestingly, higher ITH was an independent predictor of better DFS in early-stage Ns-NSCLC. Up-regulation of immune-related genes, including CRABP2, ULBP2, IL31RA, and IL1A, independently portended a dismal prognosis. Enhanced TCR diversity of peripheral blood mononuclear cells (PBMCs) predicted better prognosis, indicative of a noninvasive method for relapse surveillance. Eventually, seven machine-learning (ML) algorithms were employed to evaluate the predictive accuracy of clinical, genomic, transcriptomic, and TCR repertoire data on DFS, showing that clinical and RNA features combination in the random forest (RF) algorithm, with area under the curve (AUC) of 97.5% and 83.3% in the training and testing cohort, respectively, significantly outperformed other methods.
Conclusions: This study comprehensively profiled the genomic, transcriptomic, and TCR repertoire spectrums of Chinese early-stage Ns-NSCLC, shedding light on biological underpinnings and candidate biomarkers for prognosis development.
Keywords: Early-stage lung cancer (early-stage LC); disease-free survival (DFS); machine-learning (ML); multi-omics.
2024 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-800/coif). W.L. serves as an unpaid Associate Editor-in-Chief of Translational Lung Cancer Research from May 2023 to April 2024. Xiaoli Cui, D.W., Z.G., and H.L. are current employees of YuceBio Technology Co., Ltd. The other authors have no conflicts of interest to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
