LncRNA MIR181A2HG negatively regulates human keratinocytes proliferation by binding SRSF1
- PMID: 38736729
- PMCID: PMC11082102
- DOI: 10.1007/s10616-024-00621-6
LncRNA MIR181A2HG negatively regulates human keratinocytes proliferation by binding SRSF1
Abstract
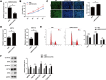
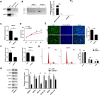
Psoriasis is a common chronic inflammatory skin disease. Abnormal proliferation of keratinocytes plays an important role in the pathogenesis of psoriasis. Long non-coding RNAs (lncRNAs) are involved in the regulation of a variety of cell biological processes. The purpose of this study was to investigate the potential role of lncRNA MIR181A2HG in the proliferation of human keratinocytes. qRT-PCR and Western blotting were performed to measure the expression levels of MIR181A2HG, SRSF1, KRT6, and KRT16 in tissue specimens and HaCaT keratinocytes. The effects of MIR181A2HG on HaCaT keratinocytes proliferation were evaluated using Cell Counting Kit-8 (CCK-8) assays, 5-Ethynyl-2'-deoxyuridine (EdU) incorporation, and cell-cycle assays. RNA pulldown-mass spectrometry (MS) was applied to identify the proteins interacting with MIR181A2HG. RNA pull-down-Western blotting and RNA immunoprecipitation coupled with real-time quantitative reverse transcription-PCR (RIP-qRT-PCR) assays were used to determine the interactions between MIR181A2HG and its RNA-binding proteins (RBPs). MIR181A2HG was down-regulated in psoriasis tissues. MIR181A2HG overexpression induced G0/G1 and G2/M phase cell cycle arrest and decreased the protein levels of KRT6, KRT16, Cyclin D1, CDK4, and Cyclin A2 in HaCaT keratinocytes. MIR181A2HG knockdown showed the opposite effect. By using RNA pulldown-MS, 356 proteins were identified to interact with MIR181A2HG potentially. Bioinformatics analysis showed that NOP56 and SRSF1 may be RNA binding proteins (RBPs) that may be interact with MIR181A2HG. Furthermore, by using RNA pull-down-Western blotting and RIP-qRT-PCR, SRSF1 was determined to interact with MIR181A2HG. Moreover, silencing of SRSF1 inhibited keratinocytes proliferation, which could be reversed with the knockdown of MIR181A2HG. Our findings indicated that MIR181A2HG can negatively regulate HaCaT keratinocytes proliferation by binding SRSF1, suggesting that MIR181A2HG and SRSF1 may serve as potential targets for the treatment of psoriasis.
Supplementary information: The online version contains supplementary material available at 10.1007/s10616-024-00621-6.
Keywords: Keratinocytes; LncRNAs; MIR181A2HG; Proliferation; Psoriasis; SRSF1.
© The Author(s), under exclusive licence to Springer Nature B.V. 2024. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

