The Asthma Risk Gene, GSDMB, Promotes Mitochondrial DNA-induced ISGs Expression
- PMID: 38737375
- PMCID: PMC11086750
- DOI: 10.35534/jrbtm.2024.10005
The Asthma Risk Gene, GSDMB, Promotes Mitochondrial DNA-induced ISGs Expression
Abstract
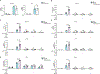
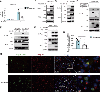
Released mitochondrial DNA (mtDNA) in cells activates cGAS-STING pathway, which induces expression of interferon-stimulated genes (ISGs) and thereby promotes inflammation, as frequently seen in asthmatic airways. However, whether the genetic determinant, Gasdermin B (GSDMB), the most replicated asthma risk gene, regulates this pathway remains unknown. We set out to determine whether and how GSDMB regulates mtDNA-activated cGAS-STING pathway and subsequent ISGs induction in human airway epithelial cells. Using qPCR, ELISA, native polyacrylamide gel electrophoresis, co-immunoprecipitation and immunofluorescence assays, we evaluated the regulation of GSDMB on cGAS-STING pathway in both BEAS-2B cells and primary normal human bronchial epithelial cells (nHBEs). mtDNA was extracted in plasma samples from human asthmatics and the correlation between mtDNA levels and eosinophil counts was analyzed. GSDMB is significantly associated with RANTES expression in asthmatic nasal epithelial brushing samples from the Genes-environments and Admixture in Latino Americans (GALA) II study. Over-expression of GSDMB promotes DNA-induced IFN and ISGs expression in bronchial epithelial BEAS-2B cells and nHBEs. Conversely, knockout of GSDMB led to weakened induction of interferon (IFNs) and ISGs in BEAS-2B cells. Mechanistically, GSDMB interacts with the C-terminus of STING, promoting the translocation of STING to Golgi, leading to the phosphorylation of IRF3 and induction of IFNs and ISGs. mtDNA copy number in serum from asthmatics was significantly correlated with blood eosinophil counts especially in male subjects. GSDMB promotes the activation of mtDNA and poly (dA:dT)-induced activation of cGAS-STING pathway in airway epithelial cells, leading to enhanced induction of ISGs.
Keywords: Airway inflammation; Asthma; GSDMB; ISGs; cGAS-STING pathway.
Conflict of interest statement
Declaration of Competing Interest A.M.K.C. is a cofounder and consultant, and equity stock holder for Proterris, which develops therapeutic uses for carbon monoxide. A.M.K.C. is a consultant and equity stock holder for SPEXIS. A.M.K.C is a member for Medforth Board of Directors. A.M.K.C. has a use patent on CO. Additionally, A.M.K.C. has a patent in COPD. Other authors declare that they have no relevant conflict of interest.
Figures


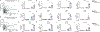
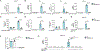
References
-
- Smit LA, Bouzigon E, Pin I, Siroux V, Monier F, Aschard H, et al. 17q21 variants modify the association between early respiratory infections and asthma. Eur. Respir. J. 2010, 36, 57–64. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
