Mitigation of TDP-43 toxic phenotype by an RGNEF fragment in amyotrophic lateral sclerosis models
- PMID: 38739752
- PMCID: PMC11146434
- DOI: 10.1093/brain/awae078
Mitigation of TDP-43 toxic phenotype by an RGNEF fragment in amyotrophic lateral sclerosis models
Abstract
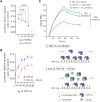
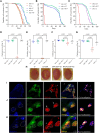
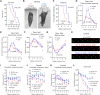
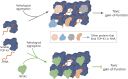
Aggregation of the RNA-binding protein TAR DNA binding protein (TDP-43) is a hallmark of TDP-proteinopathies including amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). As TDP-43 aggregation and dysregulation are causative of neuronal death, there is a special interest in targeting this protein as a therapeutic approach. Previously, we found that TDP-43 extensively co-aggregated with the dual function protein GEF (guanine exchange factor) and RNA-binding protein rho guanine nucleotide exchange factor (RGNEF) in ALS patients. Here, we show that an N-terminal fragment of RGNEF (NF242) interacts directly with the RNA recognition motifs of TDP-43 competing with RNA and that the IPT/TIG domain of NF242 is essential for this interaction. Genetic expression of NF242 in a fruit fly ALS model overexpressing TDP-43 suppressed the neuropathological phenotype increasing lifespan, abolishing motor defects and preventing neurodegeneration. Intracerebroventricular injections of AAV9/NF242 in a severe TDP-43 murine model (rNLS8) improved lifespan and motor phenotype, and decreased neuroinflammation markers. Our results demonstrate an innovative way to target TDP-43 proteinopathies using a protein fragment with a strong affinity for TDP-43 aggregates and a mechanism that includes competition with RNA sequestration, suggesting a promising therapeutic strategy for TDP-43 proteinopathies such as ALS and FTD.
Keywords: RNA binding proteins; RNA metabolism; amyotrophic lateral sclerosis; motor neuron disease; neuronal cytoplasmic inclusions; therapeutic.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures



References
-
- Strong MJ, Kesavapany S, Pant HC. The pathobiology of amyotrophic lateral sclerosis: A proteinopathy? J Neuropathol Exp Neurol. 2005;64:649–664. - PubMed
-
- Corcia P, Beltran S, Bakkouche SE, Couratier P. Therapeutic news in ALS. Rev Neurol (Paris). 2021;177:544–549. - PubMed
-
- Droppelmann CA, Campos-Melo D, Ishtiaq M, Volkening K, Strong MJ. RNA metabolism in ALS: When normal processes become pathological. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(5–6):321–336. - PubMed
-
- Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

