Microbial community structure of plant-based meat alternatives
- PMID: 38740858
- PMCID: PMC11091099
- DOI: 10.1038/s41538-024-00269-8
Microbial community structure of plant-based meat alternatives
Abstract
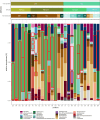
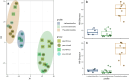
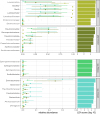
A reduction in animal-based diets has driven market demand for alternative meat products, currently raising a new generation of plant-based meat alternatives (PBMAs). It remains unclear whether these substitutes are a short-lived trend or become established in the long term. Over the last few years, the trend of increasing sales and diversifying product range has continued, but publication activities in this field are currently limited mainly to market research and food technology topics. As their popularity increases, questions emerge about the safety and nutritional risks of these novel products. Even though all the examined products must be heated before consumption, consumers lack experience with this type of product and thus further research into product safety, is desirable. To consider these issues, we examined 32 PBMAs from Austrian supermarkets. Based on 16S rRNA gene amplicon sequencing, the majority of the products were dominated by lactic acid bacteria (either Leuconostoc or Latilactobacillus), and generally had low alpha diversity. Pseudomonadota (like Pseudomonas and Shewanella) dominated the other part of the products. In addition to LABs, a high diversity of different Bacillus, but also some Enterobacteriaceae and potentially pathogenic species were isolated with the culturing approach. We assume that especially the dominance of heterofermentative LABs has high relevance for the product stability and quality with the potential to increase shelf life of the products. The number of isolated Enterobacteriaceae and potential pathogens were low, but they still demonstrated that these products are suitable for their presence.
© 2024. The Author(s).
Conflict of interest statement
The authors confirm that they have no conflicts of interest with respect to the work described in this manuscript.
Figures

References
-
- European Commission. Directorate-General for Agriculture and Rural Development. EU agricultural outlook for markets and income 2019-2030. Publications Office, 10.2762/904294 (2020).
-
- OECD. Meat consumption (indicator). 10.1787/fa290fd0-en (2022).
-
- OECD and Food and Agriculture Organization of the United Nations. OECD-FAO agricultural outlook 2022-2031. 10.1787/f1b0b29c-en (2022).
-
- Steinfeld, H. Livestock’s long shadow: Environmental issues and options, Food and Agriculture Organization of the United Nations (2006).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

