E3 ubiquitin ligase UBR5 modulates circadian rhythm by facilitating the ubiquitination and degradation of the key clock transcription factor BMAL1
- PMID: 38740904
- PMCID: PMC11336169
- DOI: 10.1038/s41401-024-01290-z
E3 ubiquitin ligase UBR5 modulates circadian rhythm by facilitating the ubiquitination and degradation of the key clock transcription factor BMAL1
Erratum in
-
Publisher Correction: E3 ubiquitin ligase UBR5 modulates circadian rhythm by facilitating the ubiquitination and degradation of the key clock transcription factor BMAL1.Acta Pharmacol Sin. 2024 Dec;45(12):2684. doi: 10.1038/s41401-024-01328-2. Acta Pharmacol Sin. 2024. PMID: 38902504 Free PMC article. No abstract available.
Abstract
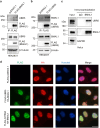
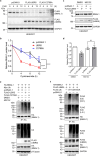
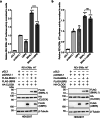
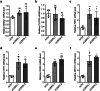
The circadian clock is the inner rhythm of life activities and is controlled by a self-sustained and endogenous molecular clock, which maintains a ~ 24 h internal oscillation. As the core element of the circadian clock, BMAL1 is susceptible to degradation through the ubiquitin-proteasome system (UPS). Nevertheless, scant information is available regarding the UPS enzymes that intricately modulate both the stability and transcriptional activity of BMAL1, affecting the cellular circadian rhythm. In this work, we identify and validate UBR5 as a new E3 ubiquitin ligase that interacts with BMAL1 by using affinity purification, mass spectrometry, and biochemical experiments. UBR5 overexpression induced BMAL1 ubiquitination, leading to diminished stability and reduced protein level of BMAL1, thereby attenuating its transcriptional activity. Consistent with this, UBR5 knockdown increases the BMAL1 protein. Domain mapping discloses that the C-terminus of BMAL1 interacts with the N-terminal domains of UBR5. Similarly, cell-line-based experiments discover that HYD, the UBR5 homolog in Drosophila, could interact with and downregulate CYCLE, the BMAL1 homolog in Drosophila. PER2-luciferase bioluminescence real-time reporting assay in a mammalian cell line and behavioral experiments in Drosophila reveal that UBR5 or hyd knockdown significantly reduces the period of the circadian clock. Therefore, our work discovers a new ubiquitin ligase UBR5 that regulates BMAL1 stability and circadian rhythm and elucidates the underlying molecular mechanism. This work provides an additional layer of complexity to the regulatory network of the circadian clock at the post-translational modification level, offering potential insights into the modulation of the dysregulated circadian rhythm.
Keywords: BMAL1; UBR5; circadian rhythm; proteomics; transcriptional activity; ubiquitination.
© 2024. The Author(s), under exclusive licence to Shanghai Institute of Materia Medica, Chinese Academy of Sciences and Chinese Pharmacological Society.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Ubiquitin ligase TRAF2 attenuates the transcriptional activity of the core clock protein BMAL1 and affects the maximal Per1 mRNA level of the circadian clock in cells.FEBS J. 2018 Aug;285(16):2987-3001. doi: 10.1111/febs.14595. Epub 2018 Jul 5. FEBS J. 2018. PMID: 29935055
-
Hyd/UBR5 defines a tumor suppressor pathway that links Polycomb repressive complex to regulated protein degradation in tissue growth control and tumorigenesis.Genes Dev. 2024 Aug 20;38(13-14):675-691. doi: 10.1101/gad.351856.124. Genes Dev. 2024. PMID: 39137945 Free PMC article.
-
Sex differences in the adrenal circadian clock: a role for BMAL1 in the regulation of urinary aldosterone excretion and renal electrolyte balance in mice.Am J Physiol Renal Physiol. 2025 Jan 1;328(1):F1-F14. doi: 10.1152/ajprenal.00177.2024. Epub 2024 Oct 24. Am J Physiol Renal Physiol. 2025. PMID: 39447118 Free PMC article.
-
Circadian Rhythm Resynchronization as a Targeted Intervention for Mood Stability in Bipolar Disorder.S D Med. 2025 May;78(suppl 5):s33. S D Med. 2025. PMID: 40550173 Review.
-
Expression of core clock genes in colorectal tumour cells compared with normal mucosa: a systematic review of clinical trials.Colorectal Dis. 2015 Apr;17(4):290-7. doi: 10.1111/codi.12847. Colorectal Dis. 2015. PMID: 25418520
Cited by
-
Nuclear p62/SQSTM1 facilitates ubiquitin-independent proteasomal degradation of BMAL1.PLoS Genet. 2025 Jul 10;21(7):e1011794. doi: 10.1371/journal.pgen.1011794. eCollection 2025 Jul. PLoS Genet. 2025. PMID: 40638681 Free PMC article.
-
Response of UBR-box E3 ubiquitin ligases and protein quality control pathways to perturbations in protein synthesis and skeletal muscle size.bioRxiv [Preprint]. 2025 Jul 27:2025.07.23.666188. doi: 10.1101/2025.07.23.666188. bioRxiv. 2025. PMID: 40777329 Free PMC article. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

