High-resolution African HLA resource uncovers HLA-DRB1 expression effects underlying vaccine response
- PMID: 38740997
- PMCID: PMC11108778
- DOI: 10.1038/s41591-024-02944-5
High-resolution African HLA resource uncovers HLA-DRB1 expression effects underlying vaccine response
Abstract
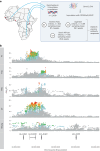
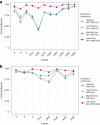
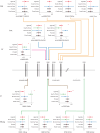
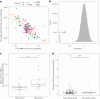
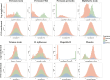
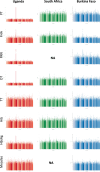
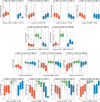
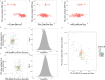
How human genetic variation contributes to vaccine effectiveness in infants is unclear, and data are limited on these relationships in populations with African ancestries. We undertook genetic analyses of vaccine antibody responses in infants from Uganda (n = 1391), Burkina Faso (n = 353) and South Africa (n = 755), identifying associations between human leukocyte antigen (HLA) and antibody response for five of eight tested antigens spanning pertussis, diphtheria and hepatitis B vaccines. In addition, through HLA typing 1,702 individuals from 11 populations of African ancestry derived predominantly from the 1000 Genomes Project, we constructed an imputation resource, fine-mapping class II HLA-DR and DQ associations explaining up to 10% of antibody response variance in our infant cohorts. We observed differences in the genetic architecture of pertussis antibody response between the cohorts with African ancestries and an independent cohort with European ancestry, but found no in silico evidence of differences in HLA peptide binding affinity or breadth. Using immune cell expression quantitative trait loci datasets derived from African-ancestry samples from the 1000 Genomes Project, we found evidence of differential HLA-DRB1 expression correlating with inferred protection from pertussis following vaccination. This work suggests that HLA-DRB1 expression may play a role in vaccine response and should be considered alongside peptide selection to improve vaccine design.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Ozawa, S. et al. Return on investment from childhood immunization in low- and middle-income countries, 2011–20. Health Aff.10.1377/hlthaff.2015.1086 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

