Unraveling transcriptomic signatures and dysregulated pathways in systemic lupus erythematosus across disease states
- PMID: 38741185
- PMCID: PMC11089778
- DOI: 10.1186/s13075-024-03327-4
Unraveling transcriptomic signatures and dysregulated pathways in systemic lupus erythematosus across disease states
Abstract
Objectives: This study aims to elucidate the transcriptomic signatures and dysregulated pathways in patients with Systemic Lupus Erythematosus (SLE), with a particular focus on those persisting during disease remission.
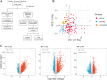
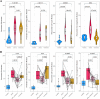
Methods: We conducted bulk RNA-sequencing of peripheral blood mononuclear cells (PBMCs) from a well-defined cohort comprising 26 remission patients meeting the Low Lupus Disease Activity State (LLDAS) criteria, 76 patients experiencing disease flares, and 15 healthy controls. To elucidate immune signature changes associated with varying disease states, we performed extensive analyses, including the identification of differentially expressed genes and pathways, as well as the construction of protein-protein interaction networks.
Results: Several transcriptomic features recovered during remission compared to the active disease state, including down-regulation of plasma and cell cycle signatures, as well as up-regulation of lymphocytes. However, specific innate immune response signatures, such as the interferon (IFN) signature, and gene modules involved in chromatin structure modification, persisted across different disease states. Drug repurposing analysis revealed certain drug classes that can target these persistent signatures, potentially preventing disease relapse.
Conclusion: Our comprehensive transcriptomic study revealed gene expression signatures for SLE in both active and remission states. The discovery of gene expression modules persisting in the remission stage may shed light on the underlying mechanisms of vulnerability to relapse in these patients, providing valuable insights for their treatment.
Keywords: Gene signatures; PBMC; Remission; Systemic lupus erythematosus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Transcriptomic features of systemic lupus erythematosus patients in flare and changes during acute in-hospital treatment.Rheumatology (Oxford). 2024 Oct 1;63(10):2810-2818. doi: 10.1093/rheumatology/kead704. Rheumatology (Oxford). 2024. PMID: 38141203
-
Single-cell RNA sequencing of peripheral blood mononuclear cells from pregnant women with Systemic lupus erythematosus.Int Rev Immunol. 2024;43(6):381-393. doi: 10.1080/08830185.2024.2376649. Epub 2024 Jul 27. Int Rev Immunol. 2024. PMID: 39066603
-
Molecular characterisation of lupus low disease activity state (LLDAS) and DORIS remission by whole-blood transcriptome-based pathways in a pan-European systemic lupus erythematosus cohort.Ann Rheum Dis. 2024 Jun 12;83(7):889-900. doi: 10.1136/ard-2023-224795. Ann Rheum Dis. 2024. PMID: 38373843 Free PMC article.
-
Transcriptomic studies unravel the molecular and cellular complexity of systemic lupus erythematosus: A review.Clin Immunol. 2024 Nov;268:110367. doi: 10.1016/j.clim.2024.110367. Epub 2024 Sep 16. Clin Immunol. 2024. PMID: 39293718 Review.
-
Microarray to deep sequencing: transcriptome and miRNA profiling to elucidate molecular pathways in systemic lupus erythematosus.Immunol Res. 2016 Feb;64(1):14-24. doi: 10.1007/s12026-015-8672-y. Immunol Res. 2016. PMID: 26188428 Review.
Cited by
-
Changes in DNA methylation are associated with systemic lupus erythematosus flare remission and clinical subtypes.Clin Epigenetics. 2024 Dec 18;16(1):181. doi: 10.1186/s13148-024-01792-x. Clin Epigenetics. 2024. PMID: 39696438 Free PMC article.
-
Single-cell transcriptional landscape of peripheral myeloid cells in autoimmune diseases.iScience. 2025 Jul 1;28(8):113026. doi: 10.1016/j.isci.2025.113026. eCollection 2025 Aug 15. iScience. 2025. PMID: 40703454 Free PMC article.
References
-
- Gilboe IM, Kvien TK, Husby G. Disease course in systemic lupus erythematosus: changes in health status, disease activity, and organ damage after 2 years. J Rheumatol. 2001;28:266–274. - PubMed
-
- Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH: Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum 1992, 35:630-640. - PubMed
MeSH terms
Grants and funding
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 17106320/General Research Fund of Hong Kong
- 2021YXNS091/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2021YXNS091/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2021YXNS091/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2023YXNS154/Jining City Science and Technology Bureau
- 2021YXNS091/Jining City Science and Technology Bureau
- ZR2019MC024/Natural Science Foundation of Shandong
- ZR2019MC024/Natural Science Foundation of Shandong
- ZR2019MC024/Natural Science Foundation of Shandong
- 07182946/Healthy and Medical Research Fund of Hong Kong
LinkOut - more resources
Full Text Sources
Medical

