The landscape of rare genetic variation associated with inflammatory bowel disease and Parkinson's disease comorbidity
- PMID: 38741190
- PMCID: PMC11092054
- DOI: 10.1186/s13073-024-01335-2
The landscape of rare genetic variation associated with inflammatory bowel disease and Parkinson's disease comorbidity
Abstract
Background: Inflammatory bowel disease (IBD) and Parkinson's disease (PD) are chronic disorders that have been suggested to share common pathophysiological processes. LRRK2 has been implicated as playing a role in both diseases. Exploring the genetic basis of the IBD-PD comorbidity through studying high-impact rare genetic variants can facilitate the identification of the novel shared genetic factors underlying this comorbidity.
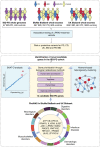
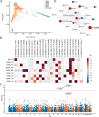
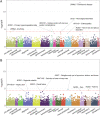
Methods: We analyzed whole exomes from the BioMe BioBank and UK Biobank, and whole genomes from a cohort of 67 European patients diagnosed with both IBD and PD to examine the effects of LRRK2 missense variants on IBD, PD and their co-occurrence (IBD-PD). We performed optimized sequence kernel association test (SKAT-O) and network-based heterogeneity clustering (NHC) analyses using high-impact rare variants in the IBD-PD cohort to identify novel candidate genes, which we further prioritized by biological relatedness approaches. We conducted phenome-wide association studies (PheWAS) employing BioMe BioBank and UK Biobank whole exomes to estimate the genetic relevance of the 14 prioritized genes to IBD-PD.
Results: The analysis of LRRK2 missense variants revealed significant associations of the G2019S and N2081D variants with IBD-PD in addition to several other variants as potential contributors to increased or decreased IBD-PD risk. SKAT-O identified two significant genes, LRRK2 and IL10RA, and NHC identified 6 significant gene clusters that are biologically relevant to IBD-PD. We observed prominent overlaps between the enriched pathways in the known IBD, PD, and candidate IBD-PD gene sets. Additionally, we detected significantly enriched pathways unique to the IBD-PD, including MAPK signaling, LPS/IL-1 mediated inhibition of RXR function, and NAD signaling. Fourteen final candidate IBD-PD genes were prioritized by biological relatedness methods. The biological importance scores estimated by protein-protein interaction networks and pathway and ontology enrichment analyses indicated the involvement of genes related to immunity, inflammation, and autophagy in IBD-PD. Additionally, PheWAS provided support for the associations of candidate genes with IBD and PD.
Conclusions: Our study confirms and uncovers new LRRK2 associations in IBD-PD. The identification of novel inflammation and autophagy-related genes supports and expands previous findings related to IBD-PD pathogenesis, and underscores the significance of therapeutic interventions for reducing systemic inflammation.
Keywords: Crohn’s disease; Genetic pleiotropy; Inflammatory bowel disease; Parkinson’s disease; Ulcerative colitis; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Lee HS, Lobbestael E, Vermeire S, Sabino J, Cleynen I. Inflammatory bowel disease and Parkinson's disease: common pathophysiological links. Gut. 2021;70(2):408–417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

