Metabolic potential of Nitrososphaera-associated clades
- PMID: 38742714
- PMCID: PMC11131427
- DOI: 10.1093/ismejo/wrae086
Metabolic potential of Nitrososphaera-associated clades
Abstract
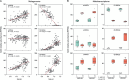
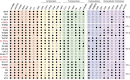
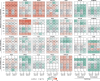
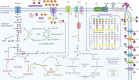
Soil ammonia-oxidizing archaea (AOA) play a crucial role in converting ammonia to nitrite, thereby mobilizing reactive nitrogen species into their soluble form, with a significant impact on nitrogen losses from terrestrial soils. Yet, our knowledge regarding their diversity and functions remains limited. In this study, we reconstructed 97 high-quality AOA metagenome-assembled genomes (MAGs) from 180 soil samples collected in Central Germany during 2014-2019 summers. These MAGs were affiliated with the order Nitrososphaerales and clustered into four family-level clades (NS-α/γ/δ/ε). Among these MAGs, 75 belonged to the most abundant but least understood δ-clade. Within the δ-clade, the amoA genes in three MAGs from neutral soils showed a 99.5% similarity to the fosmid clone 54d9, which has served as representative of the δ-clade for the past two decades since even today no cultivated representatives are available. Seventy-two MAGs constituted a distinct δ sub-clade, and their abundance and expression activity were more than twice that of other MAGs in slightly acidic soils. Unlike the less abundant clades (α, γ, and ε), the δ-MAGs possessed multiple highly expressed intracellular and extracellular carbohydrate-active enzymes responsible for carbohydrate binding (CBM32) and degradation (GH5), along with highly expressed genes involved in ammonia oxidation. Together, these results suggest metabolic versatility of uncultured soil AOA and a potential mixotrophic or chemolithoheterotrophic lifestyle among 54d9-like AOA.
Keywords: 54d9; Nitrososphaerales; ammonia-oxidizing archaea; metagenomics.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bowles TM, Atallah SS, Campbell EEet al. . Addressing agricultural nitrogen losses in a changing climate. Nat Sustainability 2018;1:399–408. 10.1038/s41893-018-0106-0 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

