Actinomycetota bioprospecting from ore-forming environments
- PMID: 38743050
- PMCID: PMC11165632
- DOI: 10.1099/mgen.0.001253
Actinomycetota bioprospecting from ore-forming environments
Abstract
Natural products from Actinomycetota have served as inspiration for many clinically relevant therapeutics. Despite early triumphs in natural product discovery, the rate of unearthing new compounds has decreased, necessitating inventive approaches. One promising strategy is to explore environments where survival is challenging. These harsh environments are hypothesized to lead to bacteria developing chemical adaptations (e.g. natural products) to enable their survival. This investigation focuses on ore-forming environments, particularly fluoride mines, which typically have extreme pH, salinity and nutrient scarcity. Herein, we have utilized metagenomics, metabolomics and evolutionary genome mining to dissect the biodiversity and metabolism in these harsh environments. This work has unveiled the promising biosynthetic potential of these bacteria and has demonstrated their ability to produce bioactive secondary metabolites. This research constitutes a pioneering endeavour in bioprospection within fluoride mining regions, providing insights into uncharted microbial ecosystems and their previously unexplored natural products.
Keywords: Actinomycetota; fluoride mines; genome mining; natural products; ore-forming environments.
Conflict of interest statement
The authors affirm that there are no conflicts of interest to disclose, ensuring complete transparency and objectivity in the research presented.
Figures
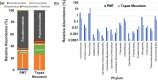

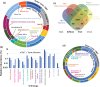


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

