The long noncoding RNA CARDINAL attenuates cardiac hypertrophy by modulating protein translation
- PMID: 38743498
- PMCID: PMC11213465
- DOI: 10.1172/JCI169112
The long noncoding RNA CARDINAL attenuates cardiac hypertrophy by modulating protein translation
Abstract
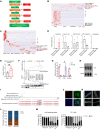
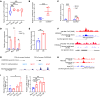
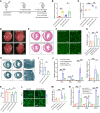
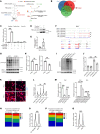
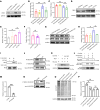
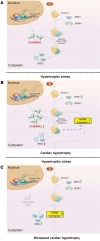
One of the features of pathological cardiac hypertrophy is enhanced translation and protein synthesis. Translational inhibition has been shown to be an effective means of treating cardiac hypertrophy, although system-wide side effects are common. Regulators of translation, such as cardiac-specific long noncoding RNAs (lncRNAs), could provide new, more targeted therapeutic approaches to inhibit cardiac hypertrophy. Therefore, we generated mice lacking a previously identified lncRNA named CARDINAL to examine its cardiac function. We demonstrate that CARDINAL is a cardiac-specific, ribosome-associated lncRNA and show that its expression was induced in the heart upon pathological cardiac hypertrophy and that its deletion in mice exacerbated stress-induced cardiac hypertrophy and augmented protein translation. In contrast, overexpression of CARDINAL attenuated cardiac hypertrophy in vivo and in vitro and suppressed hypertrophy-induced protein translation. Mechanistically, CARDINAL interacted with developmentally regulated GTP-binding protein 1 (DRG1) and blocked its interaction with DRG family regulatory protein 1 (DFRP1); as a result, DRG1 was downregulated, thereby modulating the rate of protein translation in the heart in response to stress. This study provides evidence for the therapeutic potential of targeting cardiac-specific lncRNAs to suppress disease-induced translational changes and to treat cardiac hypertrophy and heart failure.
Keywords: Cardiology; Cardiovascular disease; Development; Mouse models; Translation.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

